2C37
 
 | | RNASE PH CORE OF THE ARCHAEAL EXOSOME IN COMPLEX WITH U8 RNA | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, CHLORIDE ION, PROBABLE EXOSOME COMPLEX EXONUCLEASE 1, ... | | Authors: | Lorentzen, E, Conti, E. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of 3' End RNA Recognition and Exoribonucleolytic Cleavage by an Exosome Rnase Ph Core.
Mol.Cell, 20, 2005
|
|
2C38
 
 | | RNase PH core of the archaeal exosome in complex with A5 RNA | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, PROBABLE EXOSOME COMPLEX EXONUCLEASE 1, ... | | Authors: | Lorentzen, E, Conti, E. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of 3' End RNA Recognition and Exoribonucleolytic Cleavage by an Exosome Rnase Ph Core.
Mol.Cell, 20, 2005
|
|
8FFZ
 
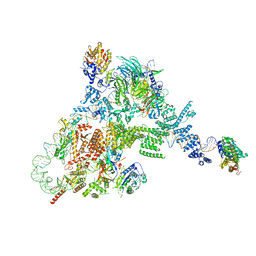 | | TFIIIA-TFIIIC-Brf1-TBP complex bound to 5S rRNA gene | | Descriptor: | DNA (151-MER), Transcription factor IIIA, Transcription factor IIIB 70 kDa subunit,TATA-box-binding protein, ... | | Authors: | Talyzina, A, He, Y. | | Deposit date: | 2022-12-11 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of TFIIIC-dependent RNA polymerase III transcription initiation.
Mol.Cell, 83, 2023
|
|
2C39
 
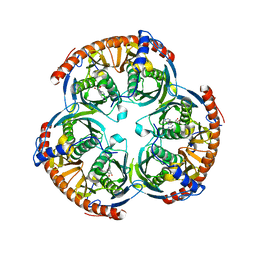 | | RNase PH core of the archaeal exosome in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PROBABLE EXOSOME COMPLEX EXONUCLEASE 1, PROBABLE EXOSOME COMPLEX EXONUCLEASE 2 | | Authors: | Lorentzen, E, Conti, E. | | Deposit date: | 2005-10-05 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of 3' End RNA Recognition and Exoribonucleolytic Cleavage by an Exosome Rnase Ph Core.
Mol.Cell, 20, 2005
|
|
5CD9
 
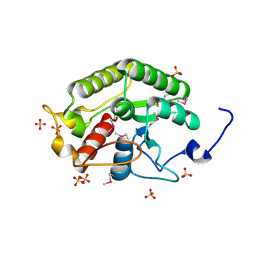 | | Crystal structure of the CTD of Drosophila Oskar protein | | Descriptor: | Maternal effect protein oskar, SULFATE ION | | Authors: | Yang, N, Hu, M, Yu, Z, Wang, M, Lehmann, R, Xu, R.M. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of Drosophila Oskar reveals a novel RNA binding protein
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8BD5
 
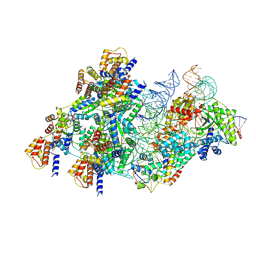 | | Cas12k-sgRNA-dsDNA-S15-TniQ-TnsC transposon recruitment complex | | Descriptor: | 30S ribosomal protein S15, ADENOSINE-5'-TRIPHOSPHATE, DNA non-target strand, ... | | Authors: | Schmitz, M, Querques, I, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
3PUF
 
 | | Crystal structure of human RNase H2 complex | | Descriptor: | Ribonuclease H2 subunit A, Ribonuclease H2 subunit B, Ribonuclease H2 subunit C | | Authors: | Figiel, M, Chon, H, Cerritelli, S.M, Cybulska, M, Crouch, R.J, Nowotny, M. | | Deposit date: | 2010-12-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi-Goutieres syndrome defects.
J.Biol.Chem., 286, 2011
|
|
6SOE
 
 | | Mouse RBM20 RRM domain | | Descriptor: | RNA-binding protein 20 | | Authors: | Mackereth, C.D, Upadhyay, S.K. | | Deposit date: | 2019-08-29 | | Release date: | 2019-11-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis of UCUU RNA motif recognition by splicing factor RBM20.
Nucleic Acids Res., 48, 2020
|
|
6MXQ
 
 | |
6QE5
 
 | | Structure of E.coli RlmJ in complex with the natural cofactor product S-adenosyl-homocysteine (SAH) | | Descriptor: | Ribosomal RNA large subunit methyltransferase J, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
5J3Y
 
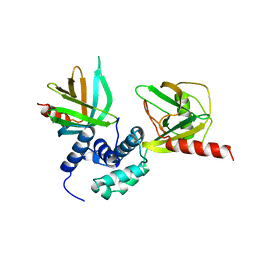 | | Crystal structure of S. pombe Dcp2:Dcp1 mRNA decapping complex | | Descriptor: | mRNA decapping complex subunit 2, mRNA-decapping enzyme subunit 1 | | Authors: | Valkov, E, Muthukumar, S, Chang, C.T, Jonas, S, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.288 Å) | | Cite: | Structure of the Dcp2-Dcp1 mRNA-decapping complex in the activated conformation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7WNU
 
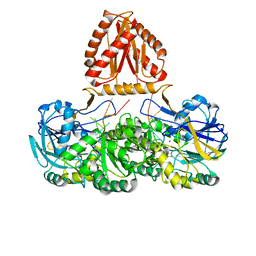 | | Mycobacterium tuberculosis Rnase J complex with 7nt RNA | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*AP*A)-3'), Ribonuclease J, ZINC ION | | Authors: | Li, J, Hu, J, Bao, L, Zhan, B, Li, Z. | | Deposit date: | 2022-01-19 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural ans functional study of RNase J from Mycobacterium tuberculosis
To Be Published
|
|
5EIM
 
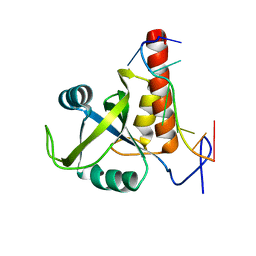 | | YTH domain-containing protein mmi1 and RNA complex | | Descriptor: | RNA (5'-R(*AP*UP*UP*AP*AP*AP*CP*A)-3'), YTH domain-containing protein mmi1 | | Authors: | Wu, B.X, Xu, J.H, Su, S.C, Ma, J.B. | | Deposit date: | 2015-10-30 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the specific recognition of DSR by the YTH domain containing protein Mmi1
Biochem. Biophys. Res. Commun., 491, 2017
|
|
5MGV
 
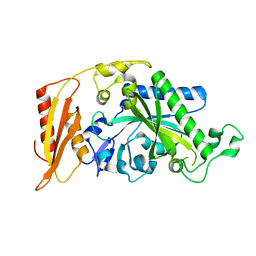 | | Kinetic and Structural Changes in HsmtPheRS, Induced by Pathogenic Mutations in Human FARS2 | | Descriptor: | Phenylalanine--tRNA ligase, mitochondrial | | Authors: | Kartvelishvili, E, Tworowski, D, Vernon, H, Chrzanowska-Lightowlers, Z, Moor, N, Wang, J, Wong, L.-J, Safro, M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Kinetic and structural changes in HsmtPheRS, induced by pathogenic mutations in human FARS2.
Protein Sci., 26, 2017
|
|
4BXZ
 
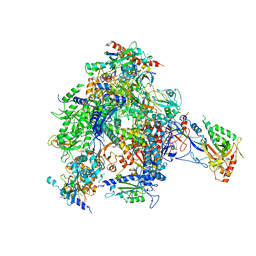 | | RNA Polymerase II-Bye1 complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-16 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6YXU
 
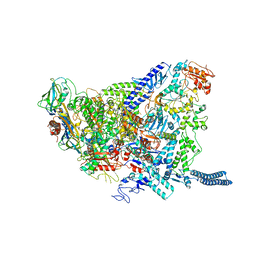 | | Structure of Mycobacterium smegmatis HelD protein in complex with RNA polymerase core - State I, primary channel engaged | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Kouba, T, Koval, T, Krasny, L, Dohnalek, J. | | Deposit date: | 2020-05-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Mycobacterial HelD is a nucleic acids-clearing factor for RNA polymerase.
Nat Commun, 11, 2020
|
|
6YYS
 
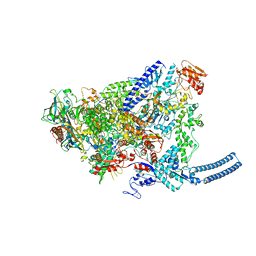 | | Structure of Mycobacterium smegmatis HelD protein in complex with RNA polymerase core - State II, primary channel engaged and active site interfering | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Kouba, T, Koval, T, Krasny, L, Dohnalek, J. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Mycobacterial HelD is a nucleic acids-clearing factor for RNA polymerase.
Nat Commun, 11, 2020
|
|
7VFT
 
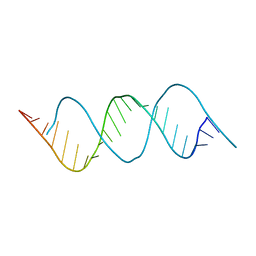 | | Crystal structure of rGGGC(CAG)5GUCC oligo | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*CP*AP*GP*CP*AP*GP*CP*AP*GP*CP*AP*GP*CP*AP*GP*GP*UP*CP*C)-3') | | Authors: | An, Y, Chan, H.Y.E, Ngo, J.C.K. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular insights into the interaction of CAG trinucleotide RNA repeats with nucleolin and its implication in polyglutamine diseases.
Nucleic Acids Res., 50, 2022
|
|
4WFX
 
 | |
5EZK
 
 | | RNA polymerase model placed by Molecular replacement into X-ray diffraction map of DNA-bound RNA Polymerase-Sigma 54 holoenzyme complex. | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darbari, V.C, Yang, Y, Lu, D, Zhang, N, Glyde, R, Wang, Y, Murakami, K.S, Buck, M, Zhang, X. | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.5 Å) | | Cite: | TRANSCRIPTION. Structures of the RNA polymerase- Sigma 54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
3ZQ4
 
 | | Unusual, dual endo- and exo-nuclease activity in the degradosome explained by crystal structure analysis of RNase J1 | | Descriptor: | CALCIUM ION, RIBONUCLEASE J 1, ZINC ION | | Authors: | Newman, J.A, Hewitt, L, Rodrigues, C, Solovyova, A, Harwood, C.R, Lewis, R.J. | | Deposit date: | 2011-06-07 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unusual, Dual Endo- and Exonuclease Activity in the Degradosome Explained by Crystal Structure Analysis of Rnase J1.
Structure, 19, 2011
|
|
6QE6
 
 | | Structure of M. capricolum TrmK in complex with the natural cofactor product S-adenosyl-homocysteine (SAH) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
3KYH
 
 | | Saccharomyces cerevisiae Cet1-Ceg1 capping apparatus | | Descriptor: | mRNA-capping enzyme subunit alpha, mRNA-capping enzyme subunit beta | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-06 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA Capping Apparatus.
Structure, 18, 2010
|
|
6SNJ
 
 | |
6STY
 
 | | Human REXO2 exonuclease in complex with RNA. | | Descriptor: | CALCIUM ION, Oligoribonuclease, mitochondrial, ... | | Authors: | Malik, D, Szewczyk, M, Szczesny, R, Nowotny, M. | | Deposit date: | 2019-09-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Human REXO2 controls short mitochondrial RNAs generated by mtRNA processing and decay machinery to prevent accumulation of double-stranded RNA.
Nucleic Acids Res., 48, 2020
|
|
