6CMO
 
 | | Rhodopsin-Gi complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab light chain, ... | | Authors: | Kang, Y, Kuybeda, O, de Waal, P.W, Mukherjee, S, Van Eps, N, Dutka, P, Zhou, X.E, Bartesaghi, A, Erramilli, S, Morizumi, T, Gu, X, Yin, Y, Liu, P, Jiang, Y, Meng, X, Zhao, G, Melcher, K, Earnst, O.P, Kossiakoff, A.A, Subramaniam, S, Xu, H.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of human rhodopsin bound to an inhibitory G protein.
Nature, 558, 2018
|
|
6B46
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF1 | | Descriptor: | Anti-CRISPR protein AcrF1, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy3, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6CRT
 
 | | Arg65Gln Mutagenic E.coli PCK | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, Phosphoenolpyruvate carboxykinase (ATP) | | Authors: | Sokaribo, A.S, Cotelesage, J.H, Novakovski, B, Goldie, H, Sanders, D. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Arg65Gln Mutagenic E.coli PCK
To Be Published
|
|
6EBM
 
 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, transmembrane domain of subunit alpha | | Descriptor: | Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6G2S
 
 | | Crystal structure of FimH in complex with a pentaflourinated biphenyl alpha D-mannoside | | Descriptor: | (2~{R},3~{S},4~{S},5~{S},6~{R})-2-(hydroxymethyl)-6-[4-[2,3,4,5,6-pentakis(fluoranyl)phenyl]phenoxy]oxane-3,4,5-triol, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin | | Authors: | Jakob, R.P, Schoenemann, W, Cramer, J, Muehlethaler, T, Daetwyler, P, Zihlmann, P, Fiege, B, Sager, C.P, Smiesko, M, Rabbani, S, Eris, D, Schwardt, O, Maier, T, Ernst, B. | | Deposit date: | 2018-03-23 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improvement of Aglycone pi-Stacking Yields Nanomolar to Sub-nanomolar FimH Antagonists.
Chemmedchem, 14, 2019
|
|
6G2R
 
 | | Crystal structure of FimH in complex with a tetraflourinated biphenyl alpha D-mannoside | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[3-chloranyl-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-phenyl]-2,3,5,6-tetrakis(fluoranyl)benzenecarbonitrile, SULFATE ION, ... | | Authors: | Jakob, R.P, Schoenemann, W, Cramer, J, Muehlethaler, T, Daetwyler, P, Zihlmann, P, Fiege, B, Sager, C.P, Smiesko, M, Rabbani, S, Eris, D, Schwardt, O, Maier, T, Ernst, B. | | Deposit date: | 2018-03-23 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improvement of Aglycone pi-Stacking Yields Nanomolar to Sub-nanomolar FimH Antagonists.
Chemmedchem, 14, 2019
|
|
8YYO
 
 | |
8YEA
 
 | |
8YYN
 
 | |
9FS9
 
 | |
9FSA
 
 | |
1TJS
 
 | | E. COLI THYMIDYLATE SYNTHASE | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1997-06-27 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
1TDU
 
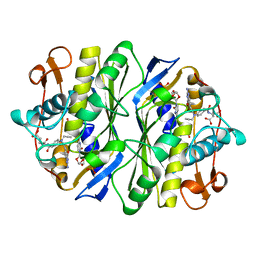 | | E. COLI THYMIDYLATE SYNTHASE IN COMPLEX WITH CB3717 AND 2'-DEOXYURIDINE (DURD) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE, PHOSPHATE ION, ... | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1997-06-25 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
1PWH
 
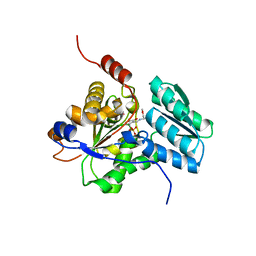 | | Rat Liver L-Serine Dehydratase- Complex with PYRIDOXYL-(O-METHYL-SERINE)-5-MONOPHOSPHATE | | Descriptor: | L-serine dehydratase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-O-METHYL-L-SERINE, POTASSIUM ION | | Authors: | Yamada, T, Komoto, J, Takata, Y, Ogawa, H, Takusagawa, F. | | Deposit date: | 2003-07-01 | | Release date: | 2003-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of serine dehydratase from rat liver.
Biochemistry, 42, 2003
|
|
1P1C
 
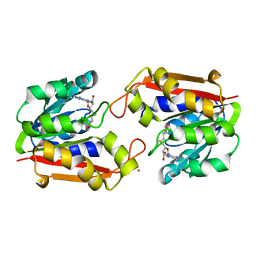 | | Guanidinoacetate Methyltransferase with Gd ion | | Descriptor: | GADOLINIUM ION, Guanidinoacetate N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Komoto, J, Takusagawa, F. | | Deposit date: | 2003-04-12 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Monoclinic guanidinoacetate methyltransferase and gadolinium ion-binding characteristics.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OZJ
 
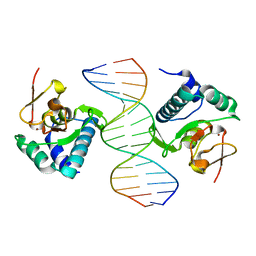 | | Crystal structure of Smad3-MH1 bound to DNA at 2.4 A resolution | | Descriptor: | SMAD 3, Smad binding element, ZINC ION | | Authors: | Chai, J, Wu, J.-W, Yan, N, Massague, J, Pavletich, N.P, Shi, Y. | | Deposit date: | 2003-04-09 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Features of a Smad3 MH1-DNA complex. Roles of water and zinc in DNA binding.
J.Biol.Chem., 278, 2003
|
|
1W7S
 
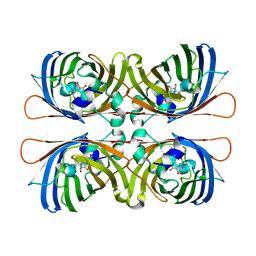 | |
1W7U
 
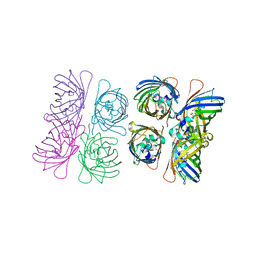 | |
1W7T
 
 | |
1PWE
 
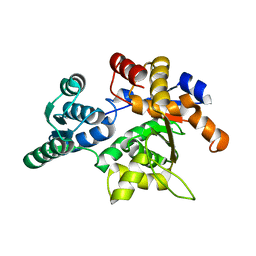 | | Rat Liver L-Serine Dehydratase Apo Enzyme | | Descriptor: | L-serine dehydratase | | Authors: | Yamada, T, Komoto, J, Takata, Y, Ogawa, H, Takusagawa, F. | | Deposit date: | 2003-07-01 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of serine dehydratase from rat liver.
Biochemistry, 42, 2003
|
|
1UXD
 
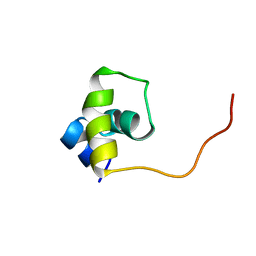 | | Fructose repressor DNA-binding domain, NMR, 34 structures | | Descriptor: | FRUCTOSE REPRESSOR | | Authors: | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | Deposit date: | 1996-12-26 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
1AAF
 
 | | NUCLEOCAPSID ZINC FINGERS DETECTED IN RETROVIRUSES: EXAFS STUDIES ON INTACT VIRUSES AND THE SOLUTION-STATE STRUCTURE OF THE NUCLEOCAPSID PROTEIN FROM HIV-1 | | Descriptor: | HIV-1 NUCLEOCAPSID PROTEIN, ZINC ION | | Authors: | Summers, M.F, Henderson, L.E, Chance, M.R, Bess Junior, J.W, South, T.L, Blake, P.R, Sagi, I, Perez-Alvarado, G, Sowder, R.C, Hare, D.R, Arthur, L.O. | | Deposit date: | 1992-04-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1.
Protein Sci., 1, 1992
|
|
1VB6
 
 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (oxygen-bound form) | | Descriptor: | Heme pas sensor protein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Watanabe, M, Sagami, I, Mikami, B, Shimizu, T. | | Deposit date: | 2004-02-24 | | Release date: | 2005-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of oxygen-bound form of a Heme PAS domain of Ec DOS
To be Published
|
|
1TRG
 
 | | E. COLI THYMIDYLATE SYNTHASE IN SYMMETRIC COMPLEX WITH CB3717 AND 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1998-05-21 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
1P1B
 
 | | Guanidinoacetate methyltransferase | | Descriptor: | Guanidinoacetate N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Komoto, J, Takusagawa, F. | | Deposit date: | 2003-04-12 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monoclinic guanidinoacetate methyltransferase and gadolinium ion-binding characteristics.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
