8HG7
 
 | | Structure of human SGLT2-MAP17 complex with Sotagliflozin | | Descriptor: | (2S,3R,4R,5S,6R)-2-[4-chloranyl-3-[(4-ethoxyphenyl)methyl]phenyl]-6-methylsulfanyl-oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, PDZK1-interacting protein 1, ... | | Authors: | Hiraizumi, M, Kishida, H, Miyaguchi, I, Nureki, O. | | Deposit date: | 2022-11-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Transport and inhibition mechanism of the human SGLT2-MAP17 glucose transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HEZ
 
 | | Structure of human SGLT2-MAP17 complex with Dapagliflozin | | Descriptor: | (2S,3R,4R,5S,6R)-2-[4-chloranyl-3-[(4-ethoxyphenyl)methyl]phenyl]-6-(hydroxymethyl)oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, PDZK1-interacting protein 1, ... | | Authors: | Hiraizumi, M, Kishida, H, Miyaguchi, I, Nureki, O. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Transport and inhibition mechanism of the human SGLT2-MAP17 glucose transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HDH
 
 | | Structure of human SGLT2-MAP17 complex with Canagliflozin | | Descriptor: | (2~{S},3~{R},4~{R},5~{S},6~{R})-2-[3-[[5-(4-fluorophenyl)thiophen-2-yl]methyl]-4-methyl-phenyl]-6-(hydroxymethyl)oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, PDZK1-interacting protein 1, ... | | Authors: | Hiraizumi, M, Kishida, H, Miyaguchi, I, Nureki, O. | | Deposit date: | 2022-11-04 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Transport and inhibition mechanism of the human SGLT2-MAP17 glucose transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6MV4
 
 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR IXa | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vadivel, K, Schreuder, H.A, Liesum, A, Bajaj, S.P. | | Deposit date: | 2018-10-24 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Sodium-site in serine protease domain of human coagulation factor IXa: evidence from the crystal structure and molecular dynamics simulations study.
J. Thromb. Haemost., 17, 2019
|
|
1K8P
 
 | |
6U02
 
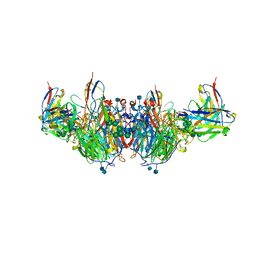 | | CryoEM-derived model of NA-63 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab-63 Heavy Chain, Fab-63 Light Chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-13 | | Release date: | 2019-12-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
4WUC
 
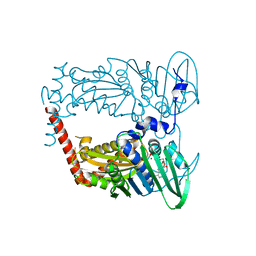 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM NaCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4X8A
 
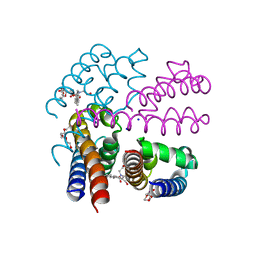 | | NavMS pore and C-terminal domain grown from protein purified in LiCl | | Descriptor: | HEGA-10, Ion transport protein, NONAETHYLENE GLYCOL, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Molecular basis of ion permeability in a voltage-gated sodium channel.
Embo J., 35, 2016
|
|
2O4F
 
 | | Structure of a parallel-stranded guanine tetraplex crystallised with monovalent ions | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*T)-3', LITHIUM ION, SODIUM ION | | Authors: | Creze, C, Rinaldi, B, Haser, R, Bouvet, P, Gouet, P. | | Deposit date: | 2006-12-04 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a d(TGGGGT) quadruplex crystallized in the presence of Li+ ions.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6A8C
 
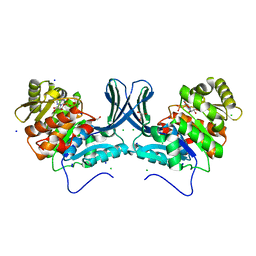 | | Ribokinase from Leishmania donovani with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gatreddi, S, Vasudevan, D, Qureshi, I.A. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
4GKH
 
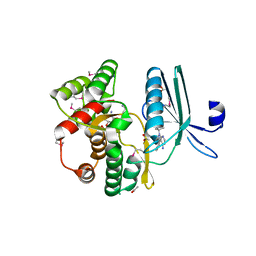 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NA-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
2NWX
 
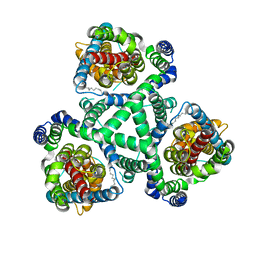 | | Crystal structure of GltPh in complex with L-aspartate and sodium ions | | Descriptor: | 425aa long hypothetical proton glutamate symport protein, ASPARTIC ACID, PALMITIC ACID, ... | | Authors: | Gouaux, E, Boudker, O, Ryan, R, Yernool, D, Shimamoto, K. | | Deposit date: | 2006-11-16 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter.
Nature, 445, 2007
|
|
6TDT
 
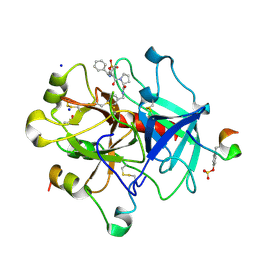 | | Thrombin in Complex with a D-DiPhe-Pro-p-pyridine derivative | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3,3-diphenyl-propanoyl]-~{N}-(pyridin-4-ylmethyl)pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2019-11-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Protein-Induced Change in Ligand Protonation during Trypsin and Thrombin Binding: Hint on Differences in Selectivity Determinants of Both Proteins?
J.Med.Chem., 63, 2020
|
|
6T4A
 
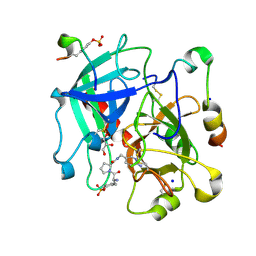 | | Thrombin in Complex with a D-Phe-Pro-p-aminopyridine derivative | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[(6-azanylpyridin-3-yl)methyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Ngo, K, Collins, C, Heine, A, Klebe, G. | | Deposit date: | 2019-10-13 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Protein-Induced Change in Ligand Protonation during Trypsin and Thrombin Binding: Hint on Differences in Selectivity Determinants of Both Proteins?
J.Med.Chem., 63, 2020
|
|
5CFY
 
 | | CRYSTAL STRUCTURE OF GLTPH R397A IN COMPLEX WITH NA+ AND L-ASP | | Descriptor: | 425aa long hypothetical proton glutamate symport protein, ASPARTIC ACID, SODIUM ION | | Authors: | Boudker, O, Oh, S. | | Deposit date: | 2015-07-08 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Coupled ion binding and structural transitions along the transport cycle of glutamate transporters.
Elife, 3, 2014
|
|
6YU7
 
 | | Crystal structure of MhsT in complex with L-tyrosine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU2
 
 | | Crystal structure of MhsT in complex with L-isoleucine | | Descriptor: | ISOLEUCINE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU3
 
 | | Crystal structure of MhsT in complex with L-phenylalanine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, PHENYLALANINE, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6L85
 
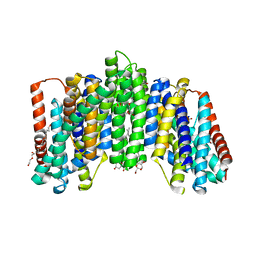 | | The sodium-dependent phosphate transporter | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Sun, Y.-J. | | Deposit date: | 2019-11-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structure of the sodium-dependent phosphate transporter reveals insights into human solute carrier SLC20.
Sci Adv, 6, 2020
|
|
6T3S
 
 | |
5ZWY
 
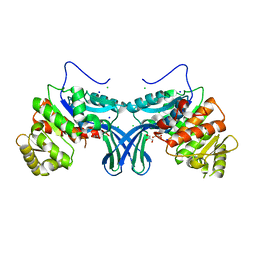 | | Ribokinase from Leishmania donovani | | Descriptor: | CHLORIDE ION, GLYCEROL, Ribokinase, ... | | Authors: | Gatreddi, S, Vasudevan, D, Qureshi, I.A. | | Deposit date: | 2018-05-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
6A8A
 
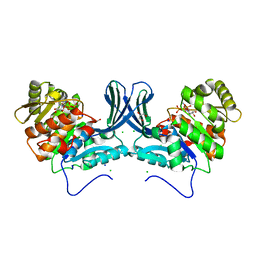 | | Ribokinase from Leishmania donovani with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gatreddi, S, Qureshi, I.A. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
6R7R
 
 | | Crystal structure of the glutamate transporter homologue GltTk in complex with D-aspartate | | Descriptor: | D-ASPARTIC ACID, DECYL-BETA-D-MALTOPYRANOSIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Arkhipova, V, Guskov, A, Slotboom, D.J. | | Deposit date: | 2019-03-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and transport of D-aspartate by the glutamate transporter homolog Glt Tk .
Elife, 8, 2019
|
|
6A8B
 
 | | Ribokinase from Leishmania donovani with AMPPCP | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Gatreddi, S, Pillalamarri, V, Qureshi, I.A. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
7UUZ
 
 | | Structure of the sodium/iodide symporter (NIS) in complex with perrhenate and sodium | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, PERRHENATE, SODIUM ION, ... | | Authors: | Ravera, S, Nicola, J.P, Salazar-De Simone, G, Sigworth, F, Karakas, E, Amzel, L.M, Bianchet, M, Carrasco, N. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the mechanism of the sodium/iodide symporter.
Nature, 612, 2022
|
|
