4IS9
 
 | | Crystal Structure of the Escherichia coli LpxC/L-161,240 complex | | Descriptor: | (4R)-2-(3,4-dimethoxy-5-propylphenyl)-N-hydroxy-4,5-dihydro-1,3-oxazole-4-carboxamide, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-01-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
7UP1
 
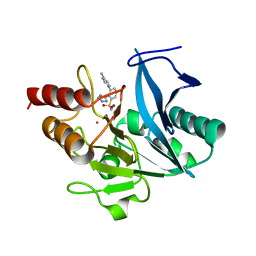 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-methyl-2-(2H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, CADMIUM ION, Metallo beta-lactamase, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7H3H
 
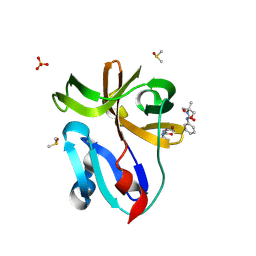 | | Group deposition for crystallographic fragment screening of Coxsackievirus A16 (G-10) 2A protease -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with Z57472297 (A71EV2A-x0395) | | Descriptor: | 1-[2-methyl-1,3-bis(oxidanyl)propan-2-yl]-3-phenyl-urea, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
8BB2
 
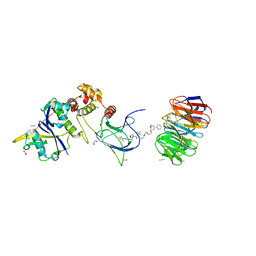 | | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with PEG linker (conformation #2) | | Descriptor: | 1,2-ETHANEDIOL, Elongin-B, Elongin-C, ... | | Authors: | Kraemer, A, Doelle, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with PEG linker (conformation #2)
To Be Published
|
|
3FMH
 
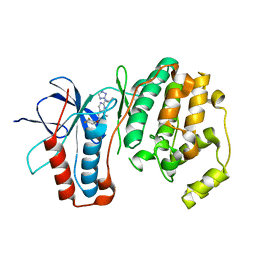 | |
7UOX
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-(hydroxymethyl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-ol, ACETATE ION, CADMIUM ION, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
8BB5
 
 | | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with Aryl linker | | Descriptor: | 1,2-ETHANEDIOL, Elongin-B, Elongin-C, ... | | Authors: | Kraemer, A, Doelle, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
5H9S
 
 | | Crystal Structure of Human Galectin-7 in Complex with TAZTDG | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, Galectin-7 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
7UP3
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (3P)-4-[4-(hydroxymethyl)phenyl]-3-(2H-tetrazol-5-yl)pyridine-2-sulfonamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
6R35
 
 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PAO1 in complex with lewis x tetrasaccharide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Lepsik, M, Sommer, R, Kuhaudomlarp, S, Lelimousin, M, Varrot, A, Titz, A, Imberty, A. | | Deposit date: | 2019-03-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Induction of rare conformation of oligosaccharide by binding to calcium-dependent bacterial lectin: X-ray crystallography and modelling study.
Eur.J.Med.Chem., 177, 2019
|
|
6GZ3
 
 | | tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis, Translocation-intermediate-POST-1 (TI-POST-1) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Flis, J, Holm, M, Rundlet, E.J, Loerke, J, Hilal, T, Dabrowski, M, Buerger, J, Mielke, T, Blanchard, S.C, Spahn, C.M.T, Budkevich, T.V. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis.
Cell Rep, 25, 2018
|
|
3IEJ
 
 | | Pyrazole-based Cathepsin S Inhibitors with Arylalkynes as P1 Binding Elements | | Descriptor: | 2-[3-{4-chloro-3-[(4-chlorophenyl)ethynyl]phenyl}-1-(3-morpholin-4-ylpropyl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl]-2-oxoethanol, Cathepsin S | | Authors: | Bembenek, S. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Pyrazole-based cathepsin S inhibitors with arylalkynes as P1 binding elements.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6GOS
 
 | | E. coli Microcin synthetase McbBCD complex with pro-MccB17 bound | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin microcin B17, CHLORIDE ION, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-04 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
9E17
 
 | | Structure of RyR1 in the primed state in the presence of caffeine (reprocessed/reanalyzed from EMPIAR-10997, 7TZC, EMD-26205) | | Descriptor: | (2S)-3-(octadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[(7-methoxy-2,3-dihydro-1,4-benzothiazepin-4(5H)-yl)methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Miotto, M.C, Marks, A.R. | | Deposit date: | 2024-10-21 | | Release date: | 2024-10-30 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Targeting ryanodine receptors with allopurinol and xanthine derivatives for the treatment of cardiac and musculoskeletal weakness disorders.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6UA1
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the no-metal bound form | | Descriptor: | 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase) | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the no-metal bound form.
To Be Published
|
|
9FOC
 
 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 11. | | Descriptor: | (2S)-1-[4-[[(3R)-1,1-bis(oxidanylidene)thiolan-3-yl]methyl-methyl-amino]-6-methyl-pyrimidin-2-yl]-N-methyl-pyrrolidine-2-carboxamide, (2S)-1-[4-[[(3S)-1,1-bis(oxidanylidene)thiolan-3-yl]methyl-methyl-amino]-6-methyl-pyrimidin-2-yl]-N-methyl-pyrrolidine-2-carboxamide, Histone-lysine N-methyltransferase NSD2 | | Authors: | Collie, G.W. | | Deposit date: | 2024-06-11 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.618 Å) | | Cite: | Structural and Molecular Insight into the PWWP1 Domain of NSD2 from the Discovery of Novel Binders Via DNA-Encoded Library Screening.
Acs Med.Chem.Lett., 16, 2025
|
|
5YG2
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS705 | | Descriptor: | 3-[1-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-fluoranyl-phenyl]piperidin-4-yl]propanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potent Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT) Featuring a Spirocyclic Scaffold
ChemMedChem, 13, 2018
|
|
1A7E
 
 | |
4BB2
 
 | | Crystal structure of cleaved corticosteroid-binding globulin in complex with progesterone | | Descriptor: | 1,2-ETHANEDIOL, CORTICOSTEROID-BINDING GLOBULIN, CYSTEINE, ... | | Authors: | Gardill, B.R, Vogl, M.R, Lin, H, Hammond, G.L, Muller, Y.A. | | Deposit date: | 2012-09-19 | | Release date: | 2012-12-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Corticosteroid-Binding Globulin: Structure-Function Implications from Species Differences
Plos One, 7, 2012
|
|
6FEB
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-1086 | | Descriptor: | (4aS,8aR)-2-[1-(2-aminoquinazolin-4-yl)piperidin-4-yl]-4-(3,4-dimethoxyphenyl)-1,2,4a,5,8,8a-hexahydrophthalazin-1-one, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2017-12-31 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-1086
To be published
|
|
5U4K
 
 | | NMR structure of the complex between the KIX domain of CBP and the transactivation domain 1 of p65 | | Descriptor: | CREB-binding protein, Transcription factor p65 | | Authors: | Lecoq, L, Raiola, L, Chabot, P.R, Cyr, N, Arseneault, G, Omichinski, J.G. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
7H2O
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z234898257 | | Descriptor: | DIMETHYL SULFOXIDE, N-methyl-1-([1,2,4]triazolo[4,3-a]pyridin-3-yl)methanamine, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
4ZUN
 
 | | Crystal structure of acetylpolyamine amidohydrolase from Mycoplana ramosa in complex with a thiol inhibitor | | Descriptor: | 5-[(3-aminopropyl)amino]pentane-1-thiol, Acetylpolyamine aminohydrolase, GLYCEROL, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2015-05-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of Their Complexes with Acetylpolyamine Amidohydrolase.
Biochemistry, 54, 2015
|
|
7H2B
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z1272517105 | | Descriptor: | DIMETHYL SULFOXIDE, Serine protease NS3, Serine protease subunit NS2B, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
4F3K
 
 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with homocysteine-DADMe-Immucillin-A | | Descriptor: | 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, {(3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-hydroxypyrrolidin-3-yl}-L-methionine | | Authors: | Haapalainen, A.M, Thomas, K, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-05-09 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with homocysteine-DADMe-Immucillin-A
Structure, 21, 2013
|
|
