3RLC
 
 | |
3R5B
 
 | |
3Q71
 
 | | Human parp14 (artd8) - macro domain 2 in complex with adenosine-5-diphosphoribose | | Descriptor: | Poly [ADP-ribose] polymerase 14, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sehic, A, Thorsell, A.G, Tresaugues, L, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
3R5D
 
 | | Pseudomonas aeruginosa DapD (PA3666) apoprotein | | Descriptor: | GLYCEROL, Tetrahydrodipicolinate N-succinyletransferase | | Authors: | Sandalova, T, Schnell, R, Schneider, G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tetrahydrodipicolinate N-succinyltransferase and dihydrodipicolinate synthase from Pseudomonas aeruginosa: structure analysis and gene deletion.
Plos One, 7, 2012
|
|
3S0Q
 
 | |
4D7Z
 
 | | E. coli L-aspartate-alpha-decarboxylase mutant N72Q to a resolution of 1.9 Angstroms | | Descriptor: | ASPARTATE 1-DECARBOXYLASE ALPHA CHAIN, ASPARTATE 1-DECARBOXYLASE BETA CHAIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bravo, J.P.K, Monteiro, D.C.F, Webb, M.E, Pearson, A.R. | | Deposit date: | 2014-12-02 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the E. Coli L-Aspartate-Alpha-Decarboxylase Mutant N72Q to a Resolution of 1.9 Angstroms
To be Published
|
|
9F19
 
 | | Human USP30 chimera in complex with NK036 inhibitor | | Descriptor: | 4-fluoranyl-~{N}-[(2~{S})-1-[[4-[(2-methyl-1-oxidanyl-propan-2-yl)sulfamoyl]phenyl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]benzamide, Ubiquitin carboxyl-terminal hydrolase 30,Ubiquitin carboxyl-terminal hydrolase 14,Ubiquitin carboxyl-terminal hydrolase 35 | | Authors: | Kazi, N.H, Gersch, M. | | Deposit date: | 2024-04-18 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Chimeric deubiquitinase engineering reveals structural basis for specific inhibition of the mitophagy regulator USP30.
Nat.Struct.Mol.Biol., 2025
|
|
9F6G
 
 | | Human USP30 chimera bound to Ubiquitin-PA | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 30,Ubiquitin carboxyl-terminal hydrolase 14,Ubiquitin carboxyl-terminal hydrolase 35, prop-2-en-1-amine | | Authors: | Kazi, N.H, Gersch, M. | | Deposit date: | 2024-05-01 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chimeric deubiquitinase engineering reveals structural basis for specific inhibition of the mitophagy regulator USP30.
Nat.Struct.Mol.Biol., 2025
|
|
1I6M
 
 | |
9EUX
 
 | |
9FBU
 
 | |
9EUZ
 
 | |
8OZB
 
 | | Crystal structure of Nup35-Nb complex | | Descriptor: | Nucleoporin NUP35, Nup35 nanobody | | Authors: | Srinivasan, V. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A checkpoint function for Nup98 in nuclear pore formation suggested by novel inhibitory nanobodies.
Embo J., 43, 2024
|
|
9F0P
 
 | | VIM-2 in complex with GKV61 (5c) - dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors | | Descriptor: | FORMIC ACID, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Bartels, K, Prester, A, Bosman, R, Gulyas, K.V, Erdelyi, M, Schulz, E.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors.
Commun Chem, 8, 2025
|
|
9F0R
 
 | | VIM-2 in complex with GKV65 (5g) - dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors | | Descriptor: | FORMIC ACID, MAGNESIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Bosman, R, Prester, A, Bartels, K, Gulyas, K.V, Erdelyi, M, Schulz, E.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors.
Commun Chem, 8, 2025
|
|
9F0Q
 
 | | VIM-2 in complex with GKV53 (5d) - dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors | | Descriptor: | FORMIC ACID, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Bartels, K, Prester, A, Bosman, R, Gulyas, K.V, Erdelyi, M, Schulz, E.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors.
Commun Chem, 8, 2025
|
|
1IGX
 
 | | Crystal Structure of Eicosapentanoic Acid Bound in the Cyclooxygenase Channel of Prostaglandin Endoperoxide H Synthase-1. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,8,11,14,17-EICOSAPENTAENOIC ACID, ... | | Authors: | Malkowski, M.G, Thuresson, E.D, Smith, W.L, Garavito, R.M. | | Deposit date: | 2001-04-18 | | Release date: | 2001-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of eicosapentaenoic and linoleic acids in the cyclooxygenase site of prostaglandin endoperoxide H synthase-1.
J.Biol.Chem., 276, 2001
|
|
9F0S
 
 | | VIM-2 in complex with GKV63 (5j) - dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors | | Descriptor: | FORMIC ACID, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Bosman, R, Prester, A, Bartels, K, Gulyas, K.V, Erdelyi, M, Schulz, E.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors.
Commun Chem, 8, 2025
|
|
4D09
 
 | | PDE2a catalytic domain in complex with a brain penetrant inhibitor | | Descriptor: | CGMP-DEPENDENT 3', 5'-CYCLIC PHOSPHODIESTERASE, MAGNESIUM ION, ... | | Authors: | Buijnsters, P, Andres, J.I, DeAngelis, M, Langlois, X, Rombouts, F, Sanderson, W, Tresadern, G, Trabanco, A, VanHoof, G, VanRoosbroeck, Y. | | Deposit date: | 2014-04-24 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of a Potent, Selective, and Brain Penetrating Pde2 Inhibitor with Demonstrated Target Engagement.
Acs Med.Chem.Lett., 5, 2014
|
|
9G8N
 
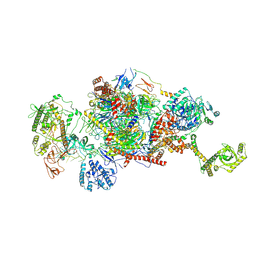 | | 80S-bound human Ski2-exosome complex | | Descriptor: | CrPV-IRES RNA, DIS3-like exonuclease 1, Exosome complex component CSL4, ... | | Authors: | Koegel, A, Keidel, A, Loukeri, M.J, Kuhn, C.C, Langer, L.M, Schaefer, I.B, Conti, E. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 635, 2024
|
|
9G8Q
 
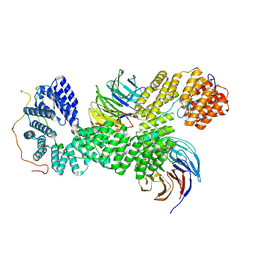 | | 40S-bound human SKI238 complex in the open state (Gatekeeping module) | | Descriptor: | Helicase SKI2W, Superkiller complex protein 3, WD repeat-containing protein 61 | | Authors: | Koegel, A, Keidel, A, Loukeri, M.J, Kuhn, C.C, Langer, L.M, Schaefer, I.B, Conti, E. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 635, 2024
|
|
9G8P
 
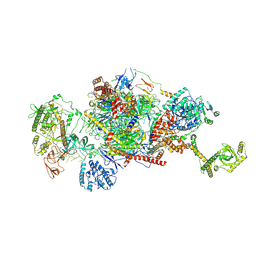 | | 40S-bound human SKI2-exosome complex | | Descriptor: | CrPV-IRES RNA, DIS3-like exonuclease 1, Exosome complex component CSL4, ... | | Authors: | Koegel, A, Keidel, A, Loukeri, M.J, Kuhn, C.C, Langer, L.M, Schaefer, I.B, Conti, E. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 635, 2024
|
|
9G8R
 
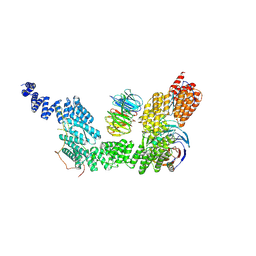 | | human SKI7-SKI238 complex in the open state | | Descriptor: | Isoform 2 of HBS1-like protein, Superkiller complex protein 2, Superkiller complex protein 3, ... | | Authors: | Koegel, A, Keidel, A, Loukeri, M.J, Kuhn, C.C, Langer, L.M, Schaefer, I.B, Conti, E. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 635, 2024
|
|
9G2J
 
 | | Thaumatin structure determined using SoS chip at ID29 (serial crystallography) | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Doak, R.B, Shoeman, R.L, Gorel, A, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2024-07-11 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Sheet-on-sheet fixed target data collection devices for serial crystallography at synchrotron and XFEL sources.
J.Appl.Crystallogr., 57, 2024
|
|
8PSM
 
 | | Asymmetric unit of the yeast fatty acid synthase in the non-rotated state with ACP at the malonyl/palmitoyl transferase domain (FASx sample) | | Descriptor: | 4'-PHOSPHOPANTETHEINE, FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
