1ADI
 
 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE AT PH 6.5 AND 25 DEGREES CELSIUS | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE | | Authors: | Silva, M.M, Poland, B.W, Hoffman, C.M, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1995-09-14 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined crystal structures of unligated adenylosuccinate synthetase from Escherichia coli.
J.Mol.Biol., 254, 1995
|
|
3TI1
 
 | | CDK2 in complex with SUNITINIB | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Alam, R, Schonbrunn, E. | | Deposit date: | 2011-08-19 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
5ZSH
 
 | | Crystal structure of monkey TLR7 in complex with CL075 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-propyl[1,3]thiazolo[4,5-c]quinolin-4-amine, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
3M9Y
 
 | | Crystal structure of Triosephosphate isomerase from methicillin resistant Staphylococcus aureus at 1.9 Angstrom resolution | | Descriptor: | CITRIC ACID, SODIUM ION, Triosephosphate isomerase | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2010-03-23 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
5ZSN
 
 | | Crystal structure of monkey TLR7 in complex with AAUUAA | | Descriptor: | 2',3'- cyclic AMP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSI
 
 | | Crystal structure of monkey TLR7 in complex with CL097 | | Descriptor: | 2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
2NMO
 
 | |
3TIZ
 
 | | CDK2 in complex with NSC 111848 | | Descriptor: | 1,2-ETHANEDIOL, 1-{(E)-[(4-hydroxyphenyl)imino]methyl}naphthalen-2-ol, Cyclin-dependent kinase 2 | | Authors: | Alam, R, Schonbrunn, E. | | Deposit date: | 2011-08-22 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
3I7E
 
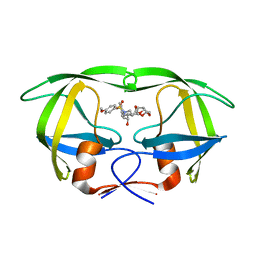 | | Co-crystal structure of HIV-1 protease bound to a mutant resistant inhibitor UIC-98038 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]CARBAMATE, HIV-1 protease | | Authors: | Hong, L, Tang, J, Ghosh, A. | | Deposit date: | 2009-07-08 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis, Protein-Ligand X-ray Structure, and Biological Evaluation of a Series of Novel Macrocyclic Human Immunodeficiency Virus-1 Protease Inhibitors to Combat Drug Resistance.
J.Med.Chem., 52, 2009
|
|
2NMN
 
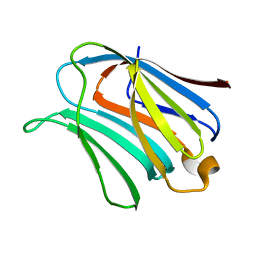 | |
3TIY
 
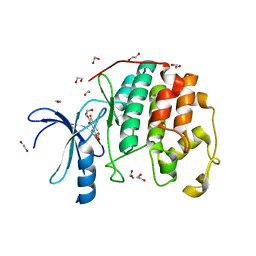 | | CDK2 in complex with NSC 35676 | | Descriptor: | 1,2-ETHANEDIOL, 2,3,4,6-tetrahydroxy-5H-benzo[7]annulen-5-one, Cyclin-dependent kinase 2 | | Authors: | Alam, R, Schonbrunn, E. | | Deposit date: | 2011-08-22 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
2NN8
 
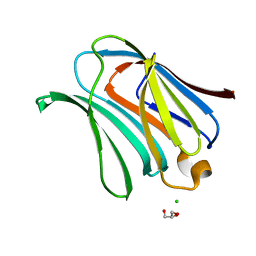 | | Crystal structure of human galectin-3 carbohydrate-recognition domain with lactose bound, at 1.35 angstrom resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, Galectin-3, ... | | Authors: | Blanchard, H, Collins, P.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Slow diffusion of lactose out of galectin-3 crystals monitored by X-ray crystallography: possible implications for ligand-exchange protocols.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1JHQ
 
 | | Three-dimensional Structure of CobT in Complex with Reaction Products of 5-methoxybenzimidazole and NaMN | | Descriptor: | N1-(5'-PHOSPHO-ALPHA-RIBOSYL)-5-METHOXYBENZIMIDAZOLE, NICOTINIC ACID, Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase | | Authors: | Cheong, C.G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
1JHY
 
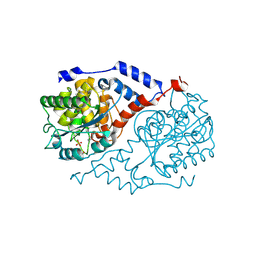 | | Three-dimensional Structure of CobT in Complex with Phenol and Nicotinate | | Descriptor: | NICOTINIC ACID, Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase, PHENOL, ... | | Authors: | Cheong, C.G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
1JHO
 
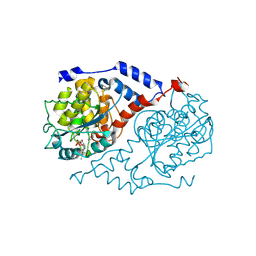 | | Three-dimensional Structure of CobT in Complex with the Reaction Products of 5-methylbenzimidazole and NaMN | | Descriptor: | N1-(5'-PHOSPHO-ALPHA-RIBOSYL)-5-METHYLBENZIMIDAZOLE, NICOTINIC ACID, Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase | | Authors: | Cheong, C.G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
1JHV
 
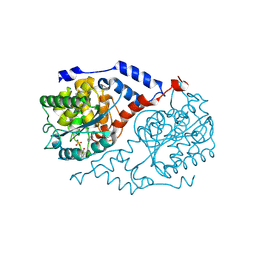 | | Three-dimensional Structure of CobT in Complex with p-cresol and Nicotinate | | Descriptor: | NICOTINIC ACID, Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase, P-CRESOL, ... | | Authors: | Cheong, C.G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
1JHM
 
 | | Three-dimensional Structure of CobT in Complex with 5-methylbenzimidazole | | Descriptor: | 5-METHYLBENZIMIDAZOLE, Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Cheong, C.G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
1JHU
 
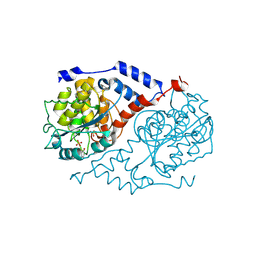 | | Three-dimensional Structure of CobT in Complex with p-cresol | | Descriptor: | Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase, P-CRESOL, PHOSPHATE ION | | Authors: | Cheong, C.G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
7AFI
 
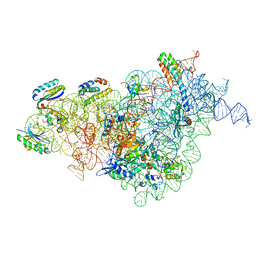 | | Bacterial 30S ribosomal subunit assembly complex state C (body domain) | | Descriptor: | 16SrRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7AF3
 
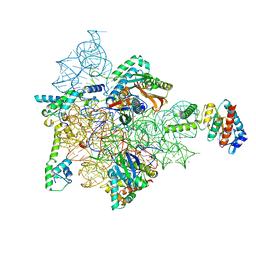 | | Bacterial 30S ribosomal subunit assembly complex state M (head domain) | | Descriptor: | 16S rRNA (head), 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S.R. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7AFN
 
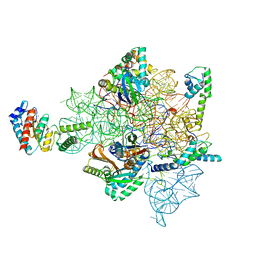 | | Bacterial 30S ribosomal subunit assembly complex state B (head domain) | | Descriptor: | 16SrRNA (head domain of the 30S ribosome), 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7AFD
 
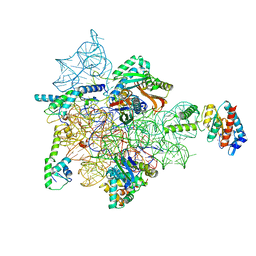 | | Bacterial 30S ribosomal subunit assembly complex state A (head domain) | | Descriptor: | 16SrRNA of the head domain (residue C931 to G1386), 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7AFL
 
 | | Bacterial 30S ribosomal subunit assembly complex state D (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16SrRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7AF8
 
 | | Bacterial 30S ribosomal subunit assembly complex state E (head domain) | | Descriptor: | 16SrRNA (head domain of the 30S ribosome, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
4W9O
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1R)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1R)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
