8D5H
 
 | | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Skarina, T, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin
To Be Published
|
|
8D5G
 
 | | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin pyrophosphate | | Descriptor: | 6-HYDROXYMETHYLPTERIN-DIPHOSPHATE, CHLORIDE ION, folP-SMZ_B27 | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin pyrophosphate
To Be Published
|
|
5KTL
 
 | |
4HQ6
 
 | | BC domain in the presence of citrate | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Heo, Y.S. | | Deposit date: | 2012-10-25 | | Release date: | 2013-10-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into the Regulation of ACC2 by Citrate
Bull.Korean Chem.Soc., 34, 2013
|
|
7JLI
 
 | | Crystal structure of Bacillus subtilis UppS | | Descriptor: | DI(HYDROXYETHYL)ETHER, Isoprenyl transferase | | Authors: | Workman, S.D, Strynadka, N.C.J. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Inhibition of Undecaprenyl Pyrophosphate Synthase from Gram-Positive Bacteria.
J.Med.Chem., 64, 2021
|
|
5MLQ
 
 | | Structure of CDPS from Nocardia brasiliensis | | Descriptor: | CDPS, CITRIC ACID | | Authors: | Bourgeois, G, Seguin, J, Moutiez, M, Babin, M, Belin, P, Mechulam, Y, Gondry, M, Schmitt, E. | | Deposit date: | 2016-12-07 | | Release date: | 2018-05-02 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structural basis for partition of the cyclodipeptide synthases into two subfamilies.
J.Struct.Biol., 203, 2018
|
|
5XNC
 
 | | Crystal structure of the branched-chain polyamine synthase (BpsA) in complex with N4-aminopropylspermidine and 5-methylthioadenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mizohata, E, Tse, K.M, Fujita, J, Inoue, T. | | Deposit date: | 2017-05-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Active site geometry of a novel aminopropyltransferase for biosynthesis of hyperthermophile-specific branched-chain polyamine.
FEBS J., 284, 2017
|
|
1RQR
 
 | | Crystal structure and mechanism of a bacterial fluorinating enzyme, product complex | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE, 5'-fluoro-5'-deoxyadenosine synthase, METHIONINE | | Authors: | Dong, C, Huang, F, Deng, H, Schaffrath, C, Spencer, J.B, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure and mechanism of a bacterial fluorinating enzyme
Nature, 427, 2004
|
|
1RQP
 
 | | Crystal structure and mechanism of a bacterial fluorinating enzyme | | Descriptor: | 5'-fluoro-5'-deoxyadenosine synthase, S-ADENOSYLMETHIONINE | | Authors: | Dong, C, Huang, F, Deng, H, Schaffrath, C, Spencer, J.B, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2003-12-06 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and mechanism of a bacterial fluorinating enzyme
Nature, 427, 2004
|
|
6ED7
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase bound to inhibitor MAC13772 | | Descriptor: | 2-[(2-nitrophenyl)sulfanyl]acetohydrazide, 7,8-diamino-pelargonic acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Brown, C.M, Zlitni, S, Chan, J, Brown, E.D, Junop, M.S. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of 7,8-diaminopelargonic acid synthase bound to inhibitor MAC13772
To Be Published
|
|
8BI9
 
 | | Structure of a cyclic beta-hairpin peptide derived from neuronal nitric oxide synthase (T112W/T116E variant) | | Descriptor: | Nitric oxide synthase, brain, TERTIARY-BUTYL ALCOHOL | | Authors: | Balboa, J.R, Ostergaard, S, Stromgaard, K, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Development of a Potent Cyclic Peptide Inhibitor of the nNOS/PSD-95 Interaction.
J.Med.Chem., 66, 2023
|
|
6KVS
 
 | | Staphylococcus aureus FabH with covalent inhibitor Oxa1 | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, 4-chloranyl-N-[[cyclopropylmethyl(methanoyl)amino]methyl]benzamide | | Authors: | Yuan, Y, Wang, J. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of S. aureus FabH, beta-ketoacyl carrier protein synthase
To Be Published
|
|
8KI5
 
 | | PhmA, a type I diterpene synthase without NST/DTE motif | | Descriptor: | (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, PhmA | | Authors: | Zhang, B, Ge, H.M, Zhu, A, Zhang, Y. | | Deposit date: | 2023-08-22 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Biosynthesis of Platelet Activating Factor Antagonist Phomactins Revealing a New Class of Type I Diterpene Synthase
To Be Published
|
|
8SK0
 
 | | Crystal structure of EvdS6 decarboxylase in ligand bound state | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sharma, P, Frigo, L, Dulin, C.C, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster.
J.Biol.Chem., 299, 2023
|
|
1ZW5
 
 | | X-ray structure of Farnesyl diphosphate synthase protein | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, ZOLEDRONIC ACID, ... | | Authors: | Kavanagh, K.L, Guo, K, Wu, X, von Delft, F, Arrowsmith, C, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-03 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular mechanism of nitrogen-containing bisphosphonates as antiosteoporosis drugs.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2H2Q
 
 | | Crystal structure of Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate synthase | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Senkovich, O, Schormann, N, Chattopadhyay, D. | | Deposit date: | 2006-05-19 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
6APM
 
 | | Hen egg-white lysozyme (WT), solved with serial millisecond crystallography using synchrotron radiation | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Lyubimov, A.Y, Mathews, I.I, Uervivojnangkoorn, M, Soltis, S.M, Cohen, A.E. | | Deposit date: | 2017-08-17 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Biochemistry, 56, 2017
|
|
4ZDO
 
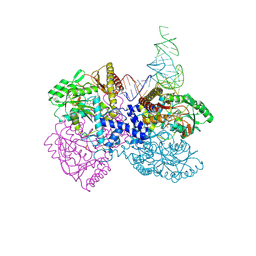 | |
4ZDP
 
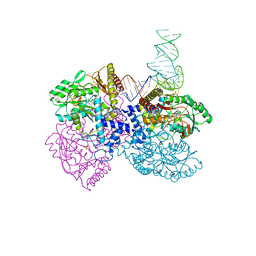 | |
5EJZ
 
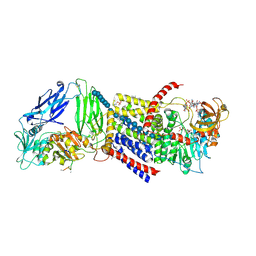 | | Bacterial Cellulose Synthase Product-Bound State | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-deoxy-2-fluoro-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ... | | Authors: | Morgan, J.L.W, Zimmer, J. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Observing cellulose biosynthesis and membrane translocation in crystallo.
Nature, 531, 2016
|
|
4I49
 
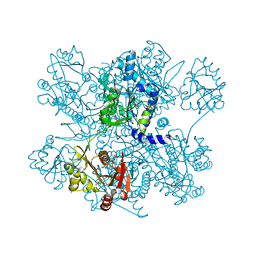 | | Structure of ngNAGS bound with bisubstrate analog CoA-NAG | | Descriptor: | (2S)-2-({(3S,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14,20-trioxo-2,4,6-trioxa-18-thia-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaicosan-20-yl}amino)pentanedioic acid (non-preferred name), Amino-acid acetyltransferase, SULFATE ION | | Authors: | Shi, D, Zhao, G, Allewell, N.M, Tuchman, M. | | Deposit date: | 2012-11-27 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the complex of Neisseria gonorrhoeae N-acetyl-l-glutamate synthase with a bound bisubstrate analog.
Biochem.Biophys.Res.Commun., 430, 2013
|
|
6OOC
 
 | | Structure of the pterocarpan synthase dirigent protein GePTS1 | | Descriptor: | Dirigent protein | | Authors: | Smith, C.A. | | Deposit date: | 2019-04-23 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pterocarpan synthase (PTS) structures suggest a common quinone methide-stabilizing function in dirigent proteins and proteins with dirigent-like domains.
J.Biol.Chem., 295, 2020
|
|
6MBH
 
 | |
4LAB
 
 | | Crystal structure of the catalytic domain of RluB | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, Ribosomal large subunit pseudouridine synthase B | | Authors: | Czudnochowski, N, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2013-06-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5043 Å) | | Cite: | The mechanism of pseudouridine synthases from a covalent complex with RNA, and alternate specificity for U2605 versus U2604 between close homologs.
Nucleic Acids Res., 42, 2014
|
|
4HP5
 
 | | The crystal structure of isomaltulose synthase mutant E295A from Erwinia rhapontici NX5 in complex with D-glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of isomaltulose synthase mutant E295A from Erwinia rhapontici NX5 in complex with D-glucose
to be published
|
|
