5EXC
 
 | |
5DRF
 
 | | Green/cyan WasCFP-pH5.5 at pH 5.5 | | Descriptor: | GLYCEROL, SODIUM ION, WasCFP-pH5.5 at pH 5.5 | | Authors: | Pletnev, V.Z, Pletneva, N.V, Pletnev, S.V. | | Deposit date: | 2015-09-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal structure of pH and T dependent green fluorescent protein WasCFP with Trp based chromophore
Russ.J.Bioorganic Chem., 42 (6), 2016
|
|
5DQB
 
 | | Green/cyan WasCFP at pH 8.0 | | Descriptor: | GLYCEROL, SODIUM ION, WasCFP_pH2 | | Authors: | Pletnev, V, Pletneva, N, Pletnev, S. | | Deposit date: | 2015-09-14 | | Release date: | 2016-07-27 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of pH and T dependent green fluorescent protein WasCFP with Trp based chromophore
Russ.J.Bioorganic Chem., 42 (6), 2016
|
|
5DQM
 
 | | Green/cyan WasCFP at pH 2.0 | | Descriptor: | WasCFP_pH2 | | Authors: | Pletnev, V.Z, Pletneva, N.V, Pletnev, S.V. | | Deposit date: | 2015-09-14 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of pH and T dependent green fluorescent protein WasCFP with Trp based chromophore
Russ.J.Bioorganic Chem., 42 (6), 2016
|
|
5DRG
 
 | | Green/cyan WasCFP at pH 10.0 | | Descriptor: | GLYCEROL, Green/cyan WasCFP_pH10 at pH 10.0, SODIUM ION | | Authors: | Pletnev, V.Z, Pletneva, N.V, Pletnev, S.V. | | Deposit date: | 2015-09-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal structure of pH and T dependent green fluorescent protein WasCFP with Trp based chromophore
Russ.J.Bioorganic Chem., 42 (6), 2016
|
|
5BT0
 
 | | Switching GFP fluorescence using genetically encoded phenyl azide chemistry through two different non-native post-translational modifications routes at the same position. | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Hartley, A.M, Worthy, H.L, Reddington, S.C, Rizkallah, P.J, Jones, D.D. | | Deposit date: | 2015-06-02 | | Release date: | 2016-07-13 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for functional switching of GFP by two disparate non-native post-translational modifications of a phenyl azide reaction handle.
Chem Sci, 7, 2016
|
|
5DY6
 
 | | Enhanced superfolder GFP with DBCO at 148 | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Jones, D.D, Rizkallah, P.J, Worthy, H.L. | | Deposit date: | 2015-09-24 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Molecular basis for functional switching of GFP by two disparate non-native post-translational modifications of a phenyl azide reaction handle.
Chem Sci, 7, 2016
|
|
5BTT
 
 | | Switching GFP fluorescence using genetically encoded phenyl azide chemistry through two different non-native post-translational modifications routes at the same position. | | Descriptor: | GLYCEROL, Green fluorescent protein, SULFATE ION | | Authors: | Hartley, A.M, Worthy, H.L, Reddington, S.C, Rizkallah, P.J, Jones, D.D. | | Deposit date: | 2015-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular basis for functional switching of GFP by two disparate non-native post-translational modifications of a phenyl azide reaction handle.
Chem Sci, 7, 2016
|
|
5BQL
 
 | | Fluorescent protein cyOFP | | Descriptor: | Fluorescent protein cyOFP | | Authors: | Sens, A, Ataie, N, Ng, H.L, Lin, M.Z, Chu, J. | | Deposit date: | 2015-05-29 | | Release date: | 2016-06-01 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A Guide to Fluorescent Protein FRET Pairs.
Sensors Basel Sensors, 16, 2016
|
|
5FVI
 
 | | Structure of IrisFP in mineral grease at 100 K. | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett., 7, 2016
|
|
5J2O
 
 | |
5FVF
 
 | | Room temperature structure of IrisFP determined by serial femtosecond crystallography. | | Descriptor: | AMMONIUM ION, Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-06 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett., 7, 2016
|
|
4Z4K
 
 | |
4Z4M
 
 | |
5FGU
 
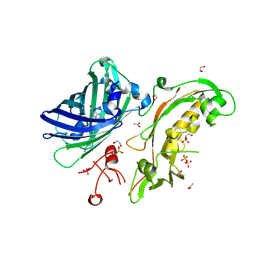 | | Structure of Sda1 nuclease apoprotein as an EGFP fixed-arm fusion | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Green fluorescent protein,Extracellular streptodornase D, ... | | Authors: | Moon, A.F, Krahn, J.M, Xun, L, Cuneo, M.J, Pedersen, L.C. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Structural characterization of the virulence factor Sda1 nuclease from Streptococcus pyogenes.
Nucleic Acids Res., 44, 2016
|
|
5AQB
 
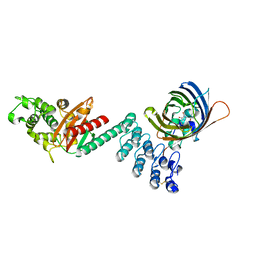 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | 3G61_DB15V4, GREEN FLUORESCENT PROTEIN | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
5HZO
 
 | | GFP mutant S205G | | Descriptor: | D-MALATE, Green fluorescent protein, UNDECYL-MALTOSIDE | | Authors: | Remington, S.J, Trujillo, K. | | Deposit date: | 2016-02-02 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Ultrafast Dynamics and Mechanisms of Proton Transfer in the GFP S205G Mutant
To Be Published
|
|
5DPJ
 
 | | sfGFP double mutant - 133/149 p-ethynyl-L-phenylalanine | | Descriptor: | Green fluorescent protein | | Authors: | Dippel, A.B, Olenginski, G.M, Maurici, N, Liskov, M.T, Brewer, S.H, Phillips-Piro, C.M. | | Deposit date: | 2015-09-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the effectiveness of spectroscopic reporter unnatural amino acids: a structural study.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5F9G
 
 | |
5DTZ
 
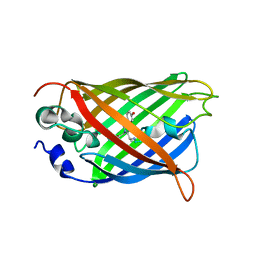 | |
5DU0
 
 | |
5H89
 
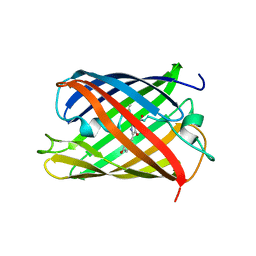 | | Crystal structure of mRojoA mutant - T16V - P63Y - W143G - L163V | | Descriptor: | mRojoA fluorescent protein | | Authors: | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
5H88
 
 | | Crystal structure of mRojoA mutant - T16V -P63F - W143A - L163V | | Descriptor: | mRojoA fluorescent protein | | Authors: | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
5H87
 
 | | Crystal structure of mRojoA mutant - P63H - W143S | | Descriptor: | mRojoA fluorescent protein | | Authors: | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-27 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
5DTY
 
 | |
