6QR1
 
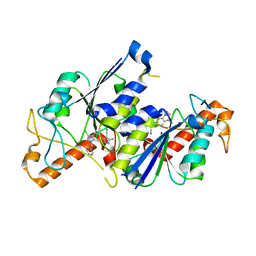 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 3-azanyl-5-[3-chloranyl-1-(pyridin-3-ylmethyl)indol-6-yl]-1~{H}-pyrazole-4-carbonitrile, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
6QRA
 
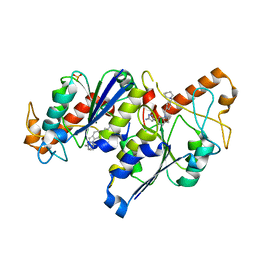 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 3-[1-[[4-(piperidin-1-ylmethyl)phenyl]methyl]indol-6-yl]-1~{H}-pyrazol-5-amine, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
9IWD
 
 | |
3NMB
 
 | |
5TPL
 
 | |
5CAP
 
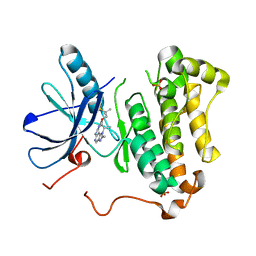 | | EGFR kinase domain mutant "TMLR" with compound 30 | | Descriptor: | 2-methyl-N-[2-(2-methyl-2-methylsulfonyl-propoxy)pyrimidin-4-yl]-1-propan-2-yl-imidazo[4,5-c]pyridin-6-amine, Epidermal growth factor receptor, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2015-06-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Noncovalent Mutant Selective Epidermal Growth Factor Receptor Inhibitors: A Lead Optimization Case Study.
J.Med.Chem., 58, 2015
|
|
2WSX
 
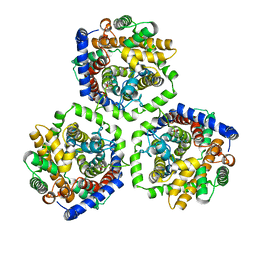 | | Crystal Structure of Carnitine Transporter from Escherichia coli | | Descriptor: | 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, L-CARNITINE/GAMMA-BUTYROBETAINE ANTIPORTER | | Authors: | Schulze, S, Terwisscha van Scheltinga, A.C, Kuehlbrandt, W. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Na(+)-Independent and Cooperative Substrate/Product Antiport in Cait.
Nature, 467, 2010
|
|
5YIJ
 
 | | Structure of a Legionella effector with substrates | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, SdeA, Ubiquitin | | Authors: | Feng, Y, Mu, Y, Wang, H. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structural basis of ubiquitin modification by the Legionella effector SdeA.
Nature, 557, 2018
|
|
5UL2
 
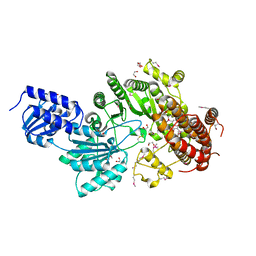 | |
4H09
 
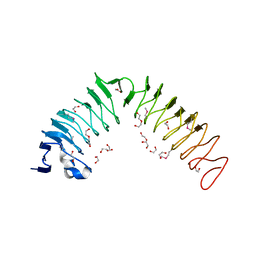 | |
7LOG
 
 | | T4 lysozyme mutant L99A in complex with 3-butylpyridine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-butylpyridine, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-10 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5KPQ
 
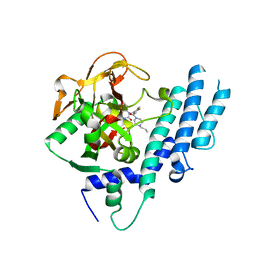 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-[(3R)-3-methyl-4-propyl-piperazin-1-yl]carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
6UBC
 
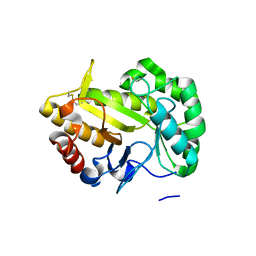 | | Crystal structure of a GH128 (subgroup VII) oligosaccharide-binding protein from Cryptococcus neoformans (CnGH128_VII) | | Descriptor: | Glyco_hydro_cc domain-containing protein | | Authors: | Santos, C.R, Costa, P.A.C.R, Souza, B.P, Vieira, P.S, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
9BP3
 
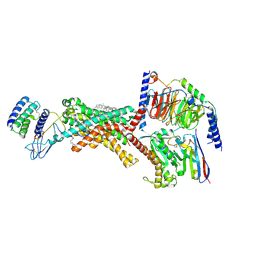 | | Human Amylin1 Receptor in complex with Gs and cagrilintide | | Descriptor: | (2S)-2-{[(1R)-1-hydroxyhexadecyl]oxy}-3-{[(1R)-1-hydroxyoctadecyl]oxy}propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Sexton, P.M, Wootten, D.L. | | Deposit date: | 2024-05-06 | | Release date: | 2025-04-23 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural and dynamic features of cagrilintide binding to calcitonin and amylin receptors.
Nat Commun, 16, 2025
|
|
5FB9
 
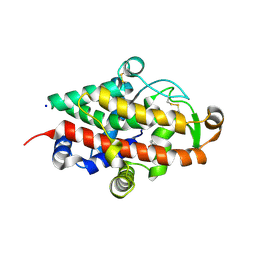 | | S1 nuclease from Aspergillus oryzae with unoccupied active site | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nuclease S1, ... | | Authors: | Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Catalytic Properties of S1 Nuclease from Aspergillus oryzae Responsible for Substrate Recognition, Cleavage, Non-Specificity, and Inhibition.
PLoS ONE, 11, 2016
|
|
6UKV
 
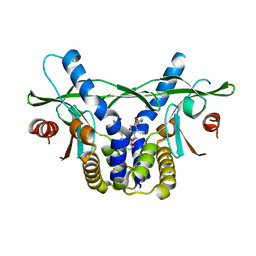 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound 9 | | Descriptor: | 4-[6-(3-{[2-(3-carboxypropanoyl)-6-methoxy-1-benzothiophen-5-yl]oxy}propoxy)-5-methoxy-1-benzothiophen-2-yl]-4-oxobutanoic acid, fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
9ITH
 
 | | Nav1.5 in complex with TTX | | Descriptor: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Wu, T. | | Deposit date: | 2024-07-20 | | Release date: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Critical role of extracellular loops in differential modulations of TTX-sensitive and TTX-resistant Na v channels.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9HFY
 
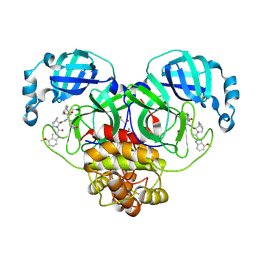 | | Crystal structure of SARS CoV-2 3CLpro (Mpro) with ALG-097078 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonyl]-~{N}-[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 1,2-ETHANEDIOL, 3C-like proteinase nsp5 | | Authors: | Tauchert, M.J, Maskos, K, McGowan, D.C, Stoycheva, A.D. | | Deposit date: | 2024-11-18 | | Release date: | 2025-07-16 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.282 Å) | | Cite: | Discovery and Preclinical Profile of ALG-097558, a Pan-Coronavirus 3CLpro Inhibitor.
J.Med.Chem., 68, 2025
|
|
6FN4
 
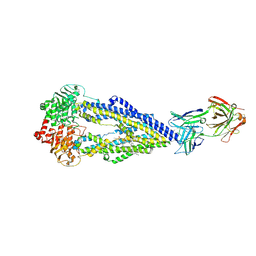 | | Apo form of UIC2 Fab complex of human-mouse chimeric ABCB1 (ABCB1HM) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Apo form of Human-mouse chimeric ABCB1 (ABCB1HM) in complex with Antigen binding fragment of UIC2., ... | | Authors: | Alam, A, Locher, K.P. | | Deposit date: | 2018-02-02 | | Release date: | 2018-02-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FQK
 
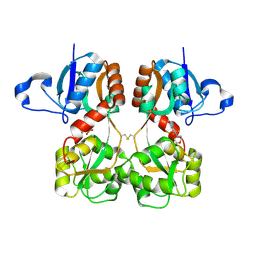 | | GluA2(flop) S729C ligand binding core dimer bound to ZK200775 at 1.98 Angstrom resolution | | Descriptor: | Glutamate receptor 2,Glutamate receptor 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98010445 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
5DMP
 
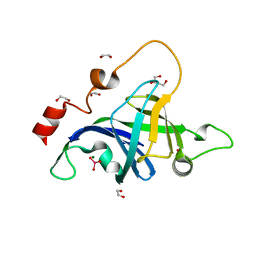 | | Structure of the Archaeal NHEJ Phosphoesterase from Methanocella paludicola. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein, ... | | Authors: | Brissett, N.C, Bartlett, E.J, Doherty, A.J. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Molecular basis for DNA strand displacement by NHEJ repair polymerases.
Nucleic Acids Res., 44, 2016
|
|
9F3A
 
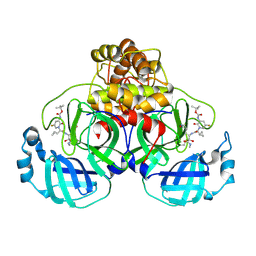 | | Crystal structure of SARS-CoV-2 Mpro in complex with RK-325 | | Descriptor: | 3C-like proteinase nsp5, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-5-fluoranyl-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
5FHN
 
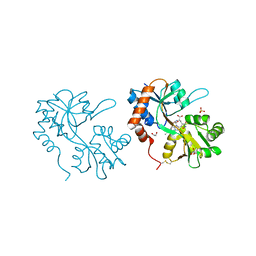 | |
9F2V
 
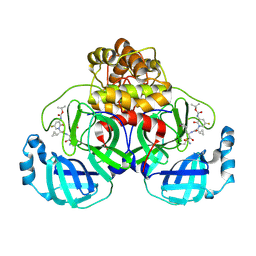 | | Crystal structure of SARS-CoV-2 Mpro in complex with RHTCR02 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-1-[[(2~{R},3~{S})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-5-fluoranyl-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-24 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
9GX8
 
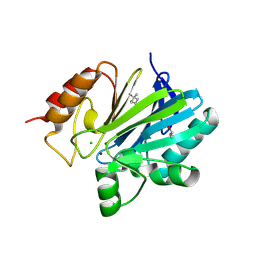 | | Crystal structure of CIM-2, a membrane-bound B1 metallo-beta-lactamase from Chryseobacterium indologenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2024-09-28 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Active site loops of membrane-anchored metallo-beta-lactamases from environmental bacteria determine cephalosporinase activity.
Antimicrob.Agents Chemother., 69, 2025
|
|
