6Q74
 
 | | PI3K delta in complex with 1benzylN[5(3,6dihydro2Hpyran4yl)2methoxypyridin3yl]2methyl1Himidazole4sulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[5-(3,6-dihydro-2~{H}-pyran-4-yl)-2-methoxy-pyridin-3-yl]-2-methyl-1-(phenylmethyl)imidazole-4-sulfonamide | | Authors: | Convery, M.A, Rowland, P, Down, K, Barton, N. | | Deposit date: | 2018-12-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of Potent, Efficient, and Selective Inhibitors of Phosphoinositide 3-Kinase delta through a Deconstruction and Regrowth Approach.
J.Med.Chem., 61, 2018
|
|
9IZO
 
 | | Apg mutant enzyme D336A of acarbose hydrolase from human gut flora K. grimontii TD1, complex with acarviosine | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-5-[[(1~{S},4~{R},5~{S},6~{S})-3-(hydroxymethyl)-4,5,6-tris(oxidanyl)cyclohex-2-en-1-yl]amino]-6-methyl-oxane-2,3,4-triol, Maltodextrin glucosidase | | Authors: | Zhou, J.H, Huang, J.Y. | | Deposit date: | 2024-08-01 | | Release date: | 2025-08-06 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Molecular insights of acarbose metabolization catalyzed by acarbose-preferred glucosidase.
Nat Commun, 16, 2025
|
|
7C2W
 
 | |
6NHR
 
 | |
5XDT
 
 | | Staphylococcus aureus FtsZ 12-316 complexed with TXA707 | | Descriptor: | 1-methylpyrrolidin-2-one, 2,6-bis(fluoranyl)-3-[[6-(trifluoromethyl)-[1,3]thiazolo[5,4-b]pyridin-2-yl]methoxy]benzamide, CALCIUM ION, ... | | Authors: | Fujita, J, Maeda, Y, Mizohata, E, Inoue, T, Kaul, M, Parhi, A.K, LaVoie, E.J, Pilch, D.S, Matsumura, H. | | Deposit date: | 2017-03-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Flexibility of an Inhibitor Overcomes Drug Resistance Mutations in Staphylococcus aureus FtsZ
ACS Chem. Biol., 12, 2017
|
|
5SIR
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1c(C)nc(c(c1)C)C(=O)Nc3nc2nc(cn2cc3)c4ccccc4, micromolar IC50=0.028279 | | Descriptor: | 3,6-dimethyl-N-[(4R)-2-phenylimidazo[1,2-a]pyrimidin-7-yl]pyridine-2-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5XFJ
 
 | | Crystal structure of LY2874455 in complex of FGFR4 gatekeeper mutation (V550M) | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | LY2874455 potently inhibits FGFR gatekeeper mutants and overcomes mutation-based resistance.
Chem. Commun. (Camb.), 54, 2018
|
|
7BTH
 
 | | Mevo lectin- Native form-1 | | Descriptor: | GLYCEROL, lectin | | Authors: | Sivaji, N, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and related studies on Mevo lectin from Methanococcus voltae A3: the first thorough characterization of an archeal lectin and its interactions.
Glycobiology, 31, 2021
|
|
7XOA
 
 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with one mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
5XG5
 
 | | Crystal structure of Mitsuba-1 with bound NAcGal | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MITSUBA-1 | | Authors: | Terada, D, Voet, A.R.D, Kamata, K, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Computational design of a symmetrical beta-trefoil lectin with cancer cell binding activity.
Sci Rep, 7, 2017
|
|
6TBG
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-(dimethylamino)-~{N}-[6-[[(1~{R},2~{R},3~{S},4~{S},5~{S})-2-(hydroxymethyl)-3,4,5-tris(oxidanyl)cyclopentyl]amino]hexyl]naphthalene-1-sulfonamide, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
7T9Q
 
 | | Crystal structure of Crocodile defensin CpoBD13 | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, CpoBD13 | | Authors: | Kvansakul, M, Williams, S, Hulett, M.D. | | Deposit date: | 2021-12-19 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crocodile defensin (CpoBD13) antifungal activity via pH-dependent phospholipid targeting and membrane disruption.
Nat Commun, 14, 2023
|
|
8V92
 
 | | BRD4 BD1 liganded with macrocyclic compound 2b (JJ-II-352A) | | Descriptor: | (4bS)-1-ethyl-21-methyl-4b,11,12,22-tetrahydro-2H,10H,18H-dibenzo[13',14':6',7'][1,5,9]trioxacyclotetradecino[12',11':4,5]pyrido[2,3-d]pyrimidine-2,4,20(1H,3H)-trione, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Schonbrunn, E, Chan, A. | | Deposit date: | 2023-12-07 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Macrocyclic dihydropyridine analogs as pan-BET BD2-preferred inhibitors.
Eur.J.Med.Chem., 290, 2025
|
|
7XO8
 
 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with three human ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7C2N
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SPIRO | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1'-(2,3-dihydro-1,4-benzodioxin-6-ylmethyl)spiro[6,7-dihydrothieno[3,2-c]pyran-4,4'-piperidine], Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-05-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
3H81
 
 | |
7ESH
 
 | | Crystal structure of amylosucrase from Calidithermus timidus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, amylosucrase | | Authors: | Tian, Y, Hou, X, Ni, D, Xu, W, Guang, C, Zhang, W, Rao, Y, Mu, W. | | Deposit date: | 2021-05-10 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based interface engineering methodology in designing a thermostable amylose-forming transglucosylase
J.Biol.Chem., 298, 2022
|
|
8DLJ
 
 | | Cryo-EM structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
5SHH
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c12c4c(ccc1[nH]c(n2)SCc3ncc(c(c3C)OC)C)ncs4, micromolar IC50=0.062527 | | Descriptor: | 7-{[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]sulfanyl}-8H-imidazo[4,5-g][1,3]benzothiazole, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Krasso, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
7XOB
 
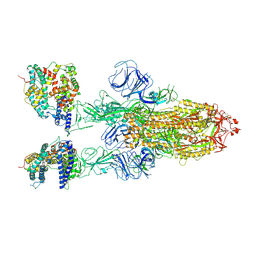 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with two mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
3HCF
 
 | |
7XIG
 
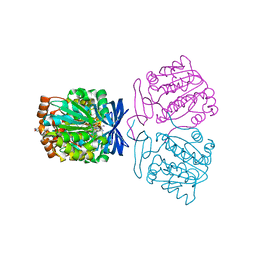 | | Crystal structure of the aminopropyltransferase, SpeE from hyperthermophilic crenarchaeon, Pyrobaculum calidifontis in complex with 5'-methylthioadenosine (MTA) and spermine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Polyamine aminopropyltransferase, ... | | Authors: | Mizohata, E, Yasuda, Y. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate Specificity of an Aminopropyltransferase and the Biosynthesis Pathway of Polyamines in the Hyperthermophilic Crenarchaeon Pyrobaculum calidifontis.
Catalysts, 12, 2022
|
|
7EUT
 
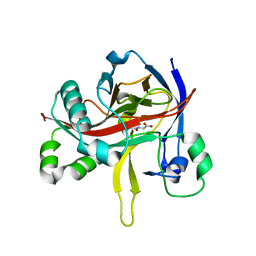 | | Crystal structures of 2-oxoglutarate dependent dioxygenase (CTB9) in complex with N-oxalylglycine | | Descriptor: | 1,2-ETHANEDIOL, 2-oxoglutarate (2-OG)-dependent dioxygenase, COPPER (II) ION, ... | | Authors: | Hou, X.D, Liu, X.Z, Yuan, Z.B, Yin, D.J, Rao, Y.J. | | Deposit date: | 2021-05-18 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Molecular Basis of the Unusual Seven-Membered Methylenedioxy Bridge Formation Catalyzed by Fe(II)/alpha-KG-Dependent Oxygenase CTB9
Acs Catalysis, 12, 2022
|
|
7EUU
 
 | | Crystal structures of 2-oxoglutarate dependent dioxygenase (CTB9) in complex with N-oxalylglycine and pre-cercosporin | | Descriptor: | 1,2-ETHANEDIOL, 2,6,11-trimethoxy-4,7,9-tris(oxidanyl)-1,12-bis[(2R)-2-oxidanylpropyl]perylene-3,10-dione, 2-oxoglutarate (2-OG)-dependent dioxygenase, ... | | Authors: | Hou, X.D, Liu, X.Z, Yuan, Z.B, Rao, Y.J. | | Deposit date: | 2021-05-18 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Molecular Basis of the Unusual Seven-Membered Methylenedioxy Bridge Formation Catalyzed by Fe(II)/alpha-KG-Dependent Oxygenase CTB9
Acs Catalysis, 12, 2022
|
|
5SK2
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1c(ccc(c1)c2nc(c(C)o2)CCOc4ncc3ccccc3c4)F, micromolar IC50=0.224356 | | Descriptor: | 3-{2-[2-(4-fluorophenyl)-5-methyl-1,3-oxazol-4-yl]ethoxy}isoquinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
