2F32
 
 | | Xray crystal structure of lysozyme mutant L20/R63A liganded to ethylguanidinium | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, N-ETHYLGUANIDINE | | Authors: | Yousef, M.S, Bischoff, N, Dyer, C.M, Baase, W.A, Matthews, B.W. | | Deposit date: | 2005-11-18 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Guanidinium derivatives bind preferentially and trigger long-distance conformational changes in an engineered T4 lysozyme.
Protein Sci., 15, 2006
|
|
2F64
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.6 A resolution with 1-METHYLQUINOLIN-2(1H)-ONE bound | | Descriptor: | 1-METHYLQUINOLIN-2(1H)-ONE, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2CVY
 
 | | Structures of Yeast Ribonucleotide Reductase I | | Descriptor: | 9-per peptide from Ribonucleoside-diphosphate reductase small chain 1, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large chain 1, ... | | Authors: | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | Deposit date: | 2005-06-14 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2CVW
 
 | | Structures of Yeast Ribonucleotide Reductase I | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large chain 1, ... | | Authors: | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | Deposit date: | 2005-06-14 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FVJ
 
 | | A novel anti-adipogenic partial agonist of peroxisome proliferator-activated receptor-gamma (PPARG) recruits pparg-coactivator-1 alpha (PGC1A) but potentiates insulin signaling in vitro | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXY-4-{[4-(2-METHOXYPHENYL)PIPERIDIN-1-YL]METHYL}ISOQUINOLINE, GLYCEROL, Nuclear receptor coactivator 1, ... | | Authors: | Benz, J, Burgermeister, E, Flament, A, Schnoebelen, A, Stihle, M, Gsell, B, Rufer, A, Ruf, A, Kuhn, B, Maerki, H.P, Mizrahi, J, Sebokova, E, Niesor, E, Meyer, M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A novel partial agonist of peroxisome proliferator-activated receptor-gamma (PPARgamma) recruits PPARgamma-coactivator-1alpha, prevents triglyceride accumulation, and potentiates insulin signaling in vitro
Mol.Endocrinol., 20, 2006
|
|
2G2O
 
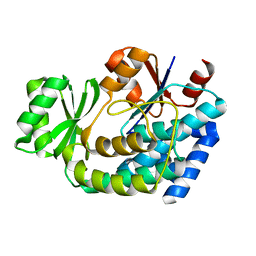 | | Structure of E.coli FabD complexed with sulfate | | Descriptor: | Malonyl CoA-acyl carrier protein transacylase, SULFATE ION | | Authors: | Oefner, C. | | Deposit date: | 2006-02-16 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Mapping the active site of Escherichia coli malonyl-CoA-acyl carrier protein transacylase (FabD) by protein crystallography.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2D7Z
 
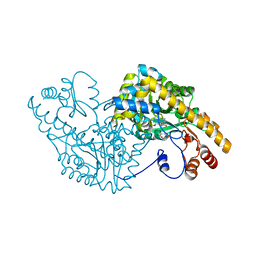 | | Aspartate Aminotransferase Mutant MAB Complexed with Maleic Acid | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-30 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D66
 
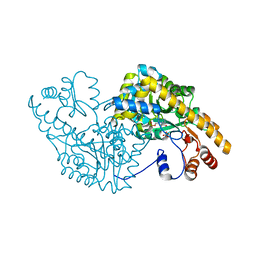 | | Aspartate Aminotransferase Mutant MAB | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
3IKD
 
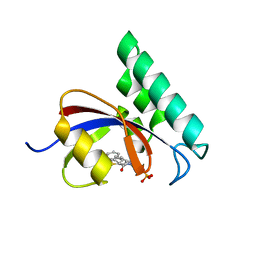 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | Descriptor: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-phenylpropyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Matthews, D, Greasley, S, Ferre, R, Parge, H. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2D65
 
 | | Aspartate Aminotransferase Mutant MABC | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
3IG8
 
 | |
3NQ8
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R4 8/5A | | Descriptor: | BENZAMIDINE, NITRATE ION, deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
1AJ2
 
 | | CRYSTAL STRUCTURE OF A BINARY COMPLEX OF E. COLI DIHYDROPTEROATE SYNTHASE | | Descriptor: | DIHYDROPTEROATE SYNTHASE, SULFATE ION, [7,8-DIHYDRO-PTERIN-6-YL METHANYL]-PHOSPHONOPHOSPHATE | | Authors: | Achari, A, Somers, D.O, Champness, J.N, Bryant, P.K, Rosemond, J, Stammers, D.K. | | Deposit date: | 1997-05-14 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase.
Nat.Struct.Biol., 4, 1997
|
|
1U5M
 
 | | Structure of a Chordin-like Cysteine-rich Repeat (VWC module) from Collagen IIA | | Descriptor: | alpha 1 type II collagen isoform 1 | | Authors: | O'Leary, J.M, Hamilton, J.m, Deane, C.M, Valeyev, N.V, Sandell, L.J, Downing, A.K. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-05 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of a prototypical chordin-like cysteine-rich repeat (von Willebrand Factor type C module) from collagen IIA
J.Biol.Chem., 279, 2004
|
|
3O05
 
 | | Crystal Structure of Yeast Pyridoxal 5-Phosphate Synthase Snz1 Complxed with Substrate PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxine biosynthesis protein SNZ1 | | Authors: | Teng, Y.B, Zhang, X, He, Y.X, Hu, H.X, Zhou, C.Z. | | Deposit date: | 2010-07-19 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the catalytic mechanism of the yeast pyridoxal 5-phosphate synthase Snz1
Biochem.J., 432, 2010
|
|
3IG5
 
 | |
1B66
 
 | | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE | | Descriptor: | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE, BIOPTERIN, ZINC ION | | Authors: | Ploom, T, Thoeny, B, Yim, J, Lee, S, Nar, H, Leimbacher, W, Huber, R, Richardson, J, Auerbach, G. | | Deposit date: | 1999-01-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and kinetic investigations on the mechanism of 6-pyruvoyl tetrahydropterin synthase.
J.Mol.Biol., 286, 1999
|
|
3O06
 
 | |
3IK8
 
 | |
3NPV
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | Descriptor: | deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
8S9M
 
 | | DNA cytosine-N4 methyltransferase (residues 79-324) from the Bdelloid rotifer Adineta vaga | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8S9N
 
 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - C2 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8S9O
 
 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - P1 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
3NQ2
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R2 3/5G | | Descriptor: | IMIDAZOLE, deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3NR0
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R6 6/10A | | Descriptor: | deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
