8P44
 
 | | Crystal structure of monkeypox virus poxin in complex with the STING agonist MD1202D | | Descriptor: | 9-[(1~{S},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, MPXVgp165 | | Authors: | Duchoslav, V, Klima, M, Boura, E. | | Deposit date: | 2023-05-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
8P45
 
 | | Crystal structure of human STING in complex with the agonist MD1202D | | Descriptor: | 9-[(1~{S},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2023-05-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
8ORW
 
 | | Crystal structure of human STING in complex with the agonist MD1203 | | Descriptor: | 9-[(1~{S},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
6RN8
 
 | | RIP2 Kinase Catalytic Domain complex with 2(4[(1,3benzothiazol5yl)amino]6(2methylpropane2sulfonyl)quinazolin7yl)oxy)ethyl phosphate | | Descriptor: | 2-[4-(1,3-benzothiazol-5-ylamino)-6-~{tert}-butylsulfonyl-quinazolin-7-yl]oxyethyl dihydrogen phosphate, CALCIUM ION, Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Convery, M.A, Haile, P.A. | | Deposit date: | 2019-05-08 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of a First-in-Class Receptor Interacting Protein 2 (RIP2) Kinase Specific Clinical Candidate, 2-((4-(Benzo[d]thiazol-5-ylamino)-6-(tert-butylsulfonyl)quinazolin-7-yl)oxy)ethyl Dihydrogen Phosphate, for the Treatment of Inflammatory Diseases.
J.Med.Chem., 62, 2019
|
|
8JBQ
 
 | |
6U5Y
 
 | |
2N89
 
 | | Tetrameric i-motif structure of dT-dC-dC-CFL-CFL-dC at acidic pH | | Descriptor: | DNA (5'-D(*TP*CP*CP*(CFL)P*(CFL)P*C)-3') | | Authors: | Abou-Assi, H, Harkness, R.W, Martin-Pintado, N, Wilds, C.J, Campos-Olivas, R, Mittermaier, A.K, Gonzalez, C, Damha, M.J. | | Deposit date: | 2015-10-09 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Stabilization of i-motif structures by 2'-beta-fluorination of DNA.
Nucleic Acids Res., 44, 2016
|
|
1A0N
 
 | | NMR STUDY OF THE SH3 DOMAIN FROM FYN PROTO-ONCOGENE TYROSINE KINASE COMPLEXED WITH THE SYNTHETIC PEPTIDE P2L CORRESPONDING TO RESIDUES 91-104 OF THE P85 SUBUNIT OF PI3-KINASE, FAMILY OF 25 STRUCTURES | | Descriptor: | FYN, PRO-PRO-ARG-PRO-LEU-PRO-VAL-ALA-PRO-GLY-SER-SER-LYS-THR | | Authors: | Renzoni, D.A, Pugh, D.J.R, Siligardi, G, Das, P, Morton, C.J, Rossi, C, Waterfield, M.D, Campbell, I.D, Ladbury, J.E. | | Deposit date: | 1997-12-05 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase.
Biochemistry, 35, 1996
|
|
6U5M
 
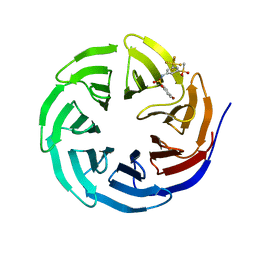 | |
6U8B
 
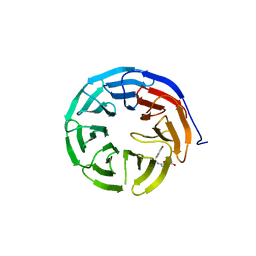 | |
6U6W
 
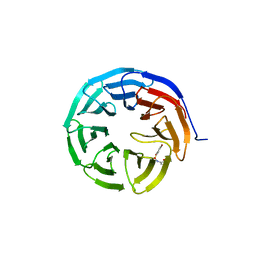 | |
7CMP
 
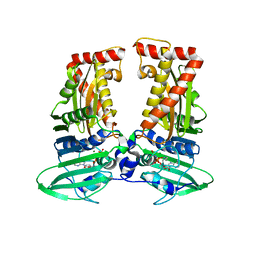 | | parE in complex with AMPPNP | | Descriptor: | DNA topoisomerase 4 subunit B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Jung, H.Y, Heo, Y.-S. | | Deposit date: | 2020-07-28 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Crystal structure of parE in complex with AMPPNP
To Be Published
|
|
6U8O
 
 | |
6U8L
 
 | |
1RH0
 
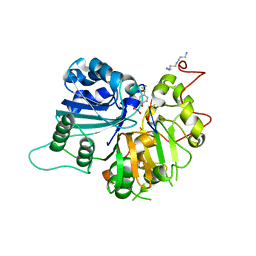 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octopamine and trinucleotide GTT | | Descriptor: | 4-(2S-AMINO-1-HYDROXYETHYL)PHENOL, 5'-D(*GP*TP*T)-3', SPERMINE, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
1T8I
 
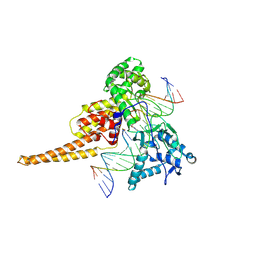 | | Human DNA Topoisomerase I (70 Kda) In Complex With The Poison Camptothecin and Covalent Complex With A 22 Base Pair DNA Duplex | | Descriptor: | 4-ETHYL-4-HYDROXY-1,12-DIHYDRO-4H-2-OXA-6,12A-DIAZA-DIBENZO[B,H]FLUORENE-3,13-DIONE, 5'-D(*(TGP)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T)-3', ... | | Authors: | Staker, B.L, Feese, M.D, Cushman, M, Pommier, Y, Zembower, D, Stewart, L, Burgin, A.B. | | Deposit date: | 2004-05-12 | | Release date: | 2005-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex
J.Med.Chem., 48, 2005
|
|
1O7C
 
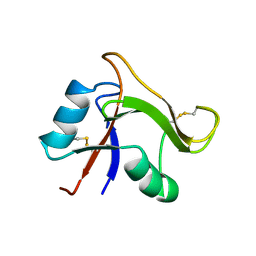 | | Solution structure of the human TSG-6 Link module in the presence of a hyaluronan octasaccharide | | Descriptor: | TUMOR NECROSIS FACTOR-INDUCIBLE PROTEIN TSG-6 | | Authors: | Blundell, C.D, Teriete, P, Kahmann, J.D, Pickford, A.R, Campbell, I.D, Day, A.J. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-23 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | The link module from ovulation- and inflammation-associated protein TSG-6 changes conformation on hyaluronan binding.
J. Biol. Chem., 278, 2003
|
|
4HKC
 
 | |
1SSZ
 
 | | Conformational Mapping of Mini-B: An N-terminal/C-terminal Construct of Surfactant Protein B Using 13C-Enhanced Fourier Transform Infrared (FTIR) Spectroscopy | | Descriptor: | Pulmonary surfactant-associated protein B | | Authors: | Waring, A.J, Walther, F.J, Gordon, L.M, Hernandez-Juviel, J.M, Hong, T, Sherman, M.A, Alonso, C, Alig, T, Braun, A, Bacon, D, Zasadzinski, J.A. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-15 | | Last modified: | 2019-04-24 | | Method: | INFRARED SPECTROSCOPY | | Cite: | The role of charged amphipathic helices in the structure and function of surfactant protein B.
J.Pept.Res., 66, 2005
|
|
2M8A
 
 | | 2'F-ANA/2'F-RNA alternated sequences | | Descriptor: | 2'F-RNA/2'F-ANA chimeric duplex | | Authors: | Martin-Pintado, N, Deleavey, G, Portella, G, Campos, R, Orozco, M, Damha, M, Gonzalez, C. | | Deposit date: | 2013-05-14 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone FCHO Hydrogen Bonds in 2'F-Substituted Nucleic Acids.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2NW7
 
 | | Crystal Structure of Tryptophan 2,3-dioxygenase (TDO) from Xanthomonas campestris in complex with ferric heme. Northeast Structural Genomics Target XcR13 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Hussain, A, Bruckmann, C, Thackray, S.J, Seetharaman, J, Tucker, T, Ho, C.K, Ma, L.C, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Chapman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OG2
 
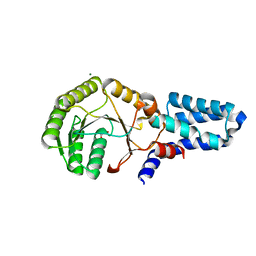 | | Crystal structure of chloroplast FtsY from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, MALONATE ION, Putative signal recognition particle receptor | | Authors: | Chartron, J, Chandrasekar, S, Ampornpan, P.J, Shan, S. | | Deposit date: | 2007-01-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Chloroplast Signal Recognition Particle (SRP) Receptor: Domain Arrangement Modulates SRP-Receptor Interaction.
J.Mol.Biol., 375, 2007
|
|
2KG4
 
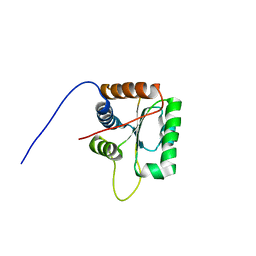 | | Three-dimensional structure of human Gadd45alpha in solution by NMR | | Descriptor: | Growth arrest and DNA-damage-inducible protein GADD45 alpha | | Authors: | Sanchez, R, Pantoja-Uceda, D, Prieto, J, Diercks, T, Campos-Olivas, R, Blanco, F.J. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human growth arrest and DNA damage 45alpha (Gadd45alpha) and its interactions with proliferating cell nuclear antigen (PCNA) and Aurora A kinase
J.Biol.Chem., 285, 2010
|
|
6UPN
 
 | | Endophilin B1 helical scaffold | | Descriptor: | Endophilin-B1 | | Authors: | Bhatt, V.S, Sundborger-Lunna, A.C. | | Deposit date: | 2019-10-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Amphipathic Motifs Regulate N-BAR Protein Endophilin B1 Auto-inhibition and Drive Membrane Remodeling.
Structure, 29, 2021
|
|
4ETC
 
 | | Lysozyme, room temperature, 24 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
