6DDR
 
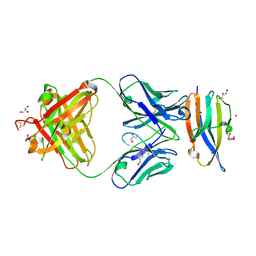 | | Crystal Structure Analysis of the Epitope of an Anti-MICA Antibody | | Descriptor: | Anti-MICA Fab fragment heavy chain clone 13A9, Anti-MICA Fab fragment light chain clone 13A9, GLYCEROL, ... | | Authors: | Matsumoto, M.L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution glycosylation site-engineering method identifies MICA epitope critical for shedding inhibition activity of anti-MICA antibodies.
MAbs, 11, 2019
|
|
6DF1
 
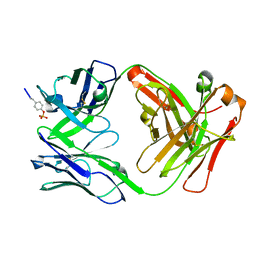 | |
6DQX
 
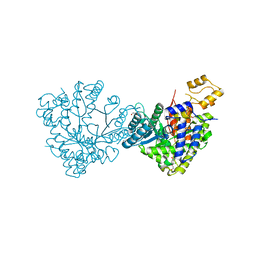 | | Actinobacillus ureae class Id ribonucleotide reductase alpha subunit | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | McBride, M.J, Palowitch, G.M, Boal, A.K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of Class Id Ribonucleotide Reductase Catalytic Subunits Reveal a Minimal Architecture for Deoxynucleotide Biosynthesis.
Biochemistry, 58, 2019
|
|
6DR3
 
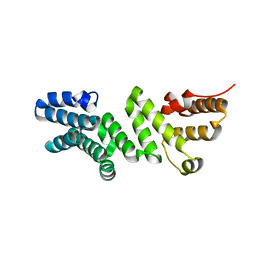 | | Crystal structure of E. coli LpoA amino terminal domain | | Descriptor: | Penicillin-binding protein activator LpoA | | Authors: | Kelley, A.C, Saper, M.A. | | Deposit date: | 2018-06-11 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal structures of the amino-terminal domain of LpoA from Escherichia coli and Haemophilus influenzae.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6DGN
 
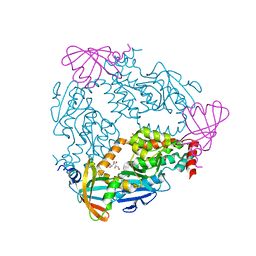 | |
6DTM
 
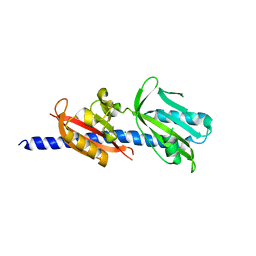 | | Crystal Structure of Helicobacter pylori TlpA Chemoreceptor Ligand Binding Domain | | Descriptor: | CHLORIDE ION, Methyl-accepting chemotaxis protein TlpA | | Authors: | Remington, S.J, Guillemin, K, Sweeney, E, Perkins, A. | | Deposit date: | 2018-06-17 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the ligand-binding domain of Helicobacter pylori chemoreceptor TlpA.
Protein Sci., 27, 2018
|
|
6DHS
 
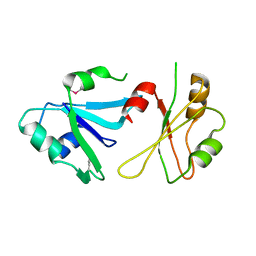 | | Structure of hnRNP H qRRM1,2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein H | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-05-21 | | Release date: | 2018-09-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Differential Conformational Dynamics Encoded by the Inter-qRRM linker of hnRNP H.
J. Am. Chem. Soc., 2018
|
|
6D4G
 
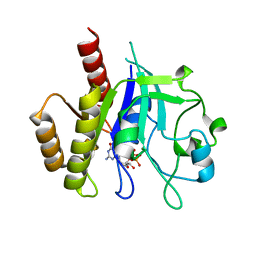 | | N-GTPase domain of p190RhoGAP-A | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rho GTPase-activating protein 35 | | Authors: | Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2018-04-18 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The N-Terminal GTPase Domain of p190RhoGAP Proteins Is a PseudoGTPase.
Structure, 26, 2018
|
|
6D6I
 
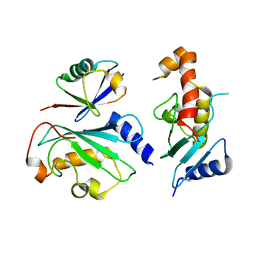 | | Ube2V1 in complex with ubiquitin variant Ubv.V1.1 and Ube2N/Ubc13 | | Descriptor: | Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 1, Ubv.V1.1 | | Authors: | Ceccarelli, D.F, Garg, P, Keszei, A, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-21 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
6D6J
 
 | |
9GHI
 
 | | Machupo virus GP1-GP2 heterodimer in complex with Fab of MAC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein G2, ... | | Authors: | Bowden, T.A, Paesen, G.C. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure and stabilization of the antigenic glycoprotein building blocks of the New World mammarenavirus spike complex.
Mbio, 16, 2025
|
|
8I9H
 
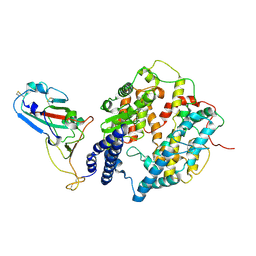 | | S-RBD (Omicron XBB.1) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9G
 
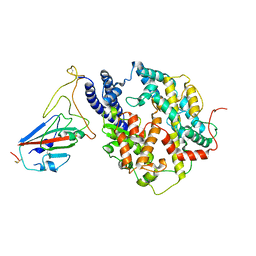 | | S-RBD (Omicron BF.7) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
6D97
 
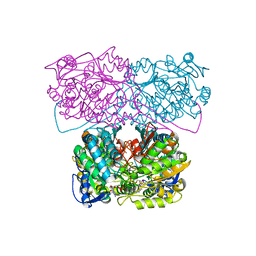 | |
6DB2
 
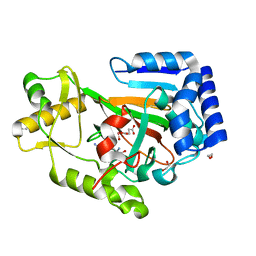 | | X-ray crystal structure of VioC bound to vanadyl ion, L-homoarginine, and succinate | | Descriptor: | 1,2-ETHANEDIOL, Alpha-ketoglutarate-dependent L-arginine hydroxylase, L-HOMOARGININE, ... | | Authors: | Dunham, N.P, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2018-05-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two Distinct Mechanisms for C-C Desaturation by Iron(II)- and 2-(Oxo)glutarate-Dependent Oxygenases: Importance of alpha-Heteroatom Assistance.
J. Am. Chem. Soc., 140, 2018
|
|
6DBB
 
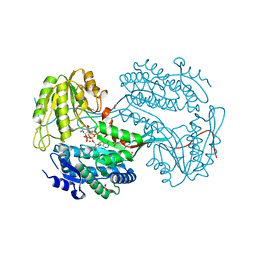 | |
8I9F
 
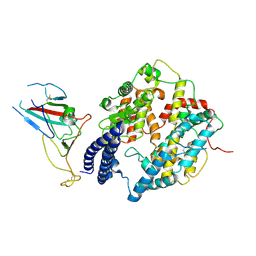 | | S-RBD (Omicron BA.2.75) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8IKX
 
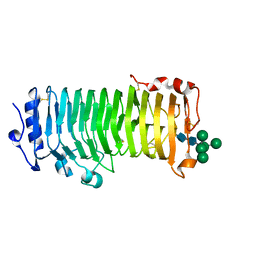 | | An Arabidopsis polygalacturonase PGLR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pectin lyase-like superfamily protein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Xiao, Y, Chai, J. | | Deposit date: | 2023-03-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A plant mechanism of hijacking pathogen virulence factors to trigger innate immunity.
Science, 383, 2024
|
|
6D88
 
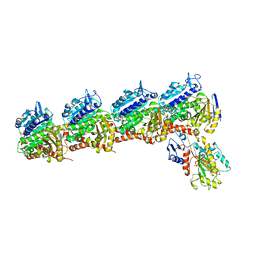 | | Tubulin-RB3_SLD-TTL in complex with compound 13f | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2018-04-26 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structural Modification of the 3,4,5-Trimethoxyphenyl Moiety in the Tubulin Inhibitor VERU-111 Leads to Improved Antiproliferative Activities.
J. Med. Chem., 61, 2018
|
|
8I9B
 
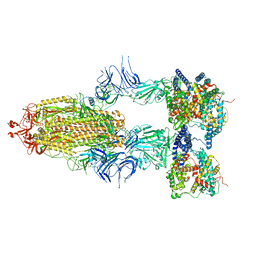 | | S-ECD (Omicron BA.2.75) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
6D9N
 
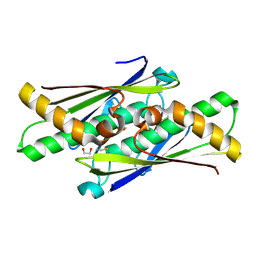 | |
6DF4
 
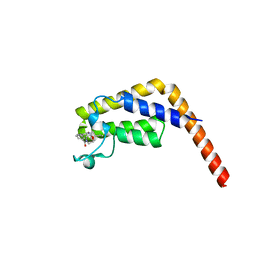 | | TAF1-BD2 in complex with Cpd8 (6-(but-3-en-1-yl)-4-(3-(morpholine-4-carbonyl)phenyl)-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one) | | Descriptor: | 6-(but-3-en-1-yl)-4-[3-(morpholine-4-carbonyl)phenyl]-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one, Transcription initiation factor TFIID subunit | | Authors: | Murray, J.M, Tang, Y. | | Deposit date: | 2018-05-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | GNE-371, a Potent and Selective Chemical Probe for the Second Bromodomains of Human Transcription-Initiation-Factor TFIID Subunit 1 and Transcription-Initiation-Factor TFIID Subunit 1-like.
J. Med. Chem., 61, 2018
|
|
6DF7
 
 | |
9GHJ
 
 | | Junin virus GP1-GP2 heterodimer in complex with Fab of JUN1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein G2, ... | | Authors: | Bowden, T.A, Paesen, G.C. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure and stabilization of the antigenic glycoprotein building blocks of the New World mammarenavirus spike complex.
Mbio, 16, 2025
|
|
6DGA
 
 | |
