5YKW
 
 | | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems | | Descriptor: | Thioredoxin-3, mitochondrial, peptide THR-PRO-VAL-CYS-THR-THR-GLU-VAL | | Authors: | Li, C.C, Yang, J, Yang, M.J, Liu, L, Peng, C.T, Li, T, He, L.H, Song, Y.J, Zhu, Y.B, Zhao, N.L, Zhao, C, Bao, R. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems
to be published
|
|
5FPU
 
 | | Crystal structure of human JARID1B in complex with GSKJ1 | | Descriptor: | 1,2-ETHANEDIOL, 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Srikannathasan, V, Nowak, R, Johansson, C, Gileadi, C, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2015-12-03 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
6C93
 
 | |
5A6Q
 
 | | Native structure of the LecB lectin from Pseudomonas aeruginosa strain PA14 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, ... | | Authors: | Sommer, R, Wagner, S, Varrot, A, Khaledi, A, Haussler, S, Imberty, A, Titz, A. | | Deposit date: | 2015-06-30 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The virulence factor LecB varies in clinical isolates: consequences for ligand binding and drug discovery.
Chem Sci, 7, 2016
|
|
5LS3
 
 | |
4TVX
 
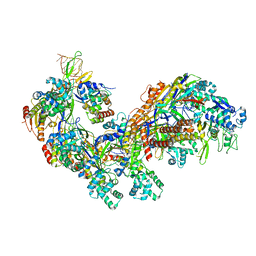 | | Crystal structure of the E. coli CRISPR RNA-guided surveillance complex, Cascade | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Jackson, R.N, Golden, S.M, Carter, J, Wiedenheft, B. | | Deposit date: | 2014-06-28 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli.
Science, 345, 2014
|
|
6RNH
 
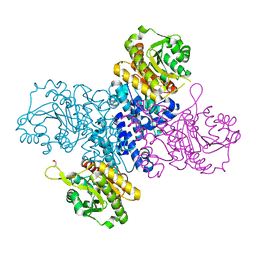 | | Structure of C-terminal truncated Plasmodium falciparum IMP-nucleotidase | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
7VYO
 
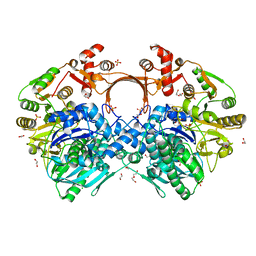 | | The structure of GdmN | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-14 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
6U3U
 
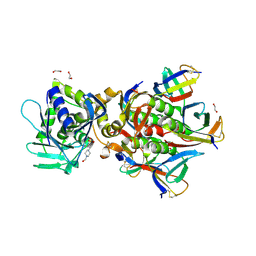 | | Crystal Structure of Shiga Toxin 2K | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Shiga toxin 2K subunit A, ... | | Authors: | Zhang, Y.Z, He, X.H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Structural and Functional Characterization of Stx2k, a New Subtype of Shiga Toxin 2.
Microorganisms, 8, 2019
|
|
4TYD
 
 | | Structure-based design of a novel series of azetidine inhibitors of the hepatitis C virus NS3/4A serine protease | | Descriptor: | (4R,6S,7Z,15S,17S)-17-[({7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)methyl]-13-methyl-N-[(1-methylcyclopropyl)sulfonyl]-2,14-dioxo-1,3,13-triazatricyclo[13.2.0.0~4,6~]heptadec-7-ene-4-carboxamide, CHLORIDE ION, NS3 protease, ... | | Authors: | Parsy, C. | | Deposit date: | 2014-07-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure-based design of a novel series of azetidine inhibitors of the hepatitis C virus NS3/4A serine protease.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6BTM
 
 | | Structure of Alternative Complex III from Flavobacterium johnsoniae (Wild Type) | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl ditetradecanoate, Alternative Complex III subunit A, Alternative Complex III subunit B, ... | | Authors: | Sun, C, Benlekbir, S, Venkatakrishnan, P, Yuhang, W, Tajkhorshid, E, Rubinstein, J.L, Gennis, R.B. | | Deposit date: | 2017-12-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the alternative complex III in a supercomplex with cytochrome oxidase.
Nature, 557, 2018
|
|
3HBG
 
 | | Structure of recombinant Chicken Liver Sulfite Oxidase mutant C185S | | Descriptor: | HYDROXY(DIOXO)MOLYBDENUM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, Sulfite Oxidase mutant C185S | | Authors: | Qiu, J.A. | | Deposit date: | 2009-05-04 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of the C185S and C185A mutants of sulfite oxidase reveal rearrangement of the active site.
Biochemistry, 49, 2010
|
|
2VZP
 
 | |
9FD4
 
 | | flavin reductase ThdF in complex with two bound FADs in space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Flavin reductase, ... | | Authors: | Bork, S, Nagel, M.F, Keller, W, Niemann, H.H. | | Deposit date: | 2024-05-16 | | Release date: | 2024-12-25 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | The NADH-dependent flavin reductase ThdF follows an ordered sequential mechanism though crystal structures reveal two FAD molecules in the active site.
J.Biol.Chem., 301, 2024
|
|
9H91
 
 | | Cryo-EM structure of the Vibrio natrigens 50S ribosomal subunit in complex with the proline-rich antimicrobial peptide Bac5(1-17). | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Raulf, K.F, Koller, T.O, Beckert, B, Morici, M, Lepak, A, Bange, G, Wilson, D.N. | | Deposit date: | 2024-10-29 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The structure of the Vibrio natriegens 70S ribosome in complex with the proline-rich antimicrobial peptide Bac5(1-17).
Nucleic Acids Res., 53, 2025
|
|
5E8X
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D,I211V,Y249F,S280T, Y282F,S287N,A350C,L352F) IN COMPLEX WITH STAUROSPORINE | | Descriptor: | GLYCEROL, STAUROSPORINE, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of apo and inhibitor-bound TGF beta R2 kinase domain: insights into TGF beta R isoform selectivity.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7VCF
 
 | | Cryo-EM structure of Chlamydomonas TOC-TIC supercomplex | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, J, Yan, Z, Jin, Z, Zhang, Y. | | Deposit date: | 2021-09-02 | | Release date: | 2022-11-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of a TOC-TIC supercomplex spanning two chloroplast envelope membranes.
Cell, 185, 2022
|
|
9H6V
 
 | | Lotus japonicus CERK6 extracellular domain in complex with two nanobodies | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gysel, K, Alsarraf, H.M.A.B, Hansen, S.B, Andersen, K.R. | | Deposit date: | 2024-10-25 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis for size-selective perception of chitin in plants
To Be Published
|
|
8SQ0
 
 | | Cleaved Ycf1p Dimer in the IFwide-beta conformation | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, Metal resistance protein YCF1, ... | | Authors: | Bickers, S.C, Benlekbir, S, Rubinstein, J.L, Kanelis, V. | | Deposit date: | 2023-05-04 | | Release date: | 2024-05-08 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a dimeric full-length ABC transporter.
Nat Commun, 15, 2024
|
|
8SV1
 
 | | Caspase-1 complex with interleukin-18 | | Descriptor: | Caspase-1, Interleukin-18 | | Authors: | Dong, Y, Pascal, D, Jon, K, Wu, H. | | Deposit date: | 2023-05-15 | | Release date: | 2024-05-08 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural transitions enable interleukin-18 maturation and signaling.
Immunity, 57, 2024
|
|
3H7I
 
 | | Structure of the metal-free D132N T4 RNase H | | Descriptor: | Ribonuclease H, SULFATE ION | | Authors: | Tomanicek, S.J, Mueser, T.C. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Additional Order Appears in the Absence of Metals in a FEN-1 protein: Structural Analysis of Magnesium Binding to Bacteriophage T4 RNaseH
To be Published
|
|
1NC7
 
 | | Crystal Structure of Thermotoga maritima 1070 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Analysis of Thermotoga maritima Hypothetical protein TM1070
To be Published
|
|
7US1
 
 | | Structure of parkin (R0RB) bound to two phospho-ubiquitin molecules | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin, ... | | Authors: | Fakih, R, Sauve, V, Gehring, K. | | Deposit date: | 2022-04-22 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Structure of the second phosphoubiquitin-binding site in parkin.
J.Biol.Chem., 298, 2022
|
|
6FMJ
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-Acetamidopropanethioyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl) benzyl)pyrrolidine-2-carboxamide (ligand 3) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-acetamidopropanethioyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Soares, P, Lucas, X, Ciulli, A. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Thioamide substitution to probe the hydroxyproline recognition of VHL ligands.
Bioorg. Med. Chem., 26, 2018
|
|
9HGC
 
 | |
