4UXN
 
 | | LSD1(KDM1A)-CoREST in complex with Z-Pro derivative of MC2580 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, ... | | Authors: | Rodriguez, V, Valente, S, Stazi, G, Lucidi, A, Mercurio, C, Vianello, P, Ciossani, G, Mattevi, A, Botrugno, O.A, Dessanti, P, Minucci, S, Varasi, M, Mai, A. | | Deposit date: | 2014-08-27 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Pyrrole- and Indole-Containing Tranylcypromine Derivatives as Novel Lysine-Specific Demethylase 1 Inhibitors Active on Cancer Cells
Chemmedchem, 6, 2015
|
|
7RRO
 
 | | Structure of the 48-nm repeat doublet microtubule from bovine tracheal cilia | | Descriptor: | Armadillo repeat containing 4, Chromosome 3 C1orf194 homolog, Cilia and flagella associated protein 161, ... | | Authors: | Gui, M, Anderson, J.R, Botsch, J.J, Meleppattu, S, Singh, S.K, Zhang, Q, Brown, A. | | Deposit date: | 2021-08-10 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | De novo identification of mammalian ciliary motility proteins using cryo-EM.
Cell, 184, 2021
|
|
9MGN
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and compound 41 | | Descriptor: | 1,2-ETHANEDIOL, 2-(cyclobutylamino)-N-[(2S)-2-hydroxy-3-{6-[(1H-pyrazol-4-yl)methoxy]-3,4-dihydroisoquinolin-2(1H)-yl}propyl]pyridine-4-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2024-12-11 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | MTA-Cooperative PRMT5 Inhibitors: Mechanism Switching Through Structure-Based Design.
J.Med.Chem., 68, 2025
|
|
9MGM
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and compound 24 | | Descriptor: | 2-(cyclobutylamino)-N-[(2S)-2-hydroxy-3-{6-[(1-methyl-1H-pyrazol-5-yl)methoxy]-3,4-dihydroisoquinolin-2(1H)-yl}propyl]pyridine-4-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2024-12-11 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | MTA-Cooperative PRMT5 Inhibitors: Mechanism Switching Through Structure-Based Design.
J.Med.Chem., 68, 2025
|
|
9MGQ
 
 | | Crystal structure of PRMT5:MEP50 in complex with sinefungin and compound 47 | | Descriptor: | 1,2-ETHANEDIOL, 2-(cyclobutylamino)-N-{(2R)-2-hydroxy-2-[(3S)-1,2,3,4-tetrahydroisoquinolin-3-yl]ethyl}pyridine-4-carboxamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2024-12-11 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | MTA-Cooperative PRMT5 Inhibitors: Mechanism Switching Through Structure-Based Design.
J.Med.Chem., 68, 2025
|
|
9MGL
 
 | |
9MGP
 
 | |
9MGR
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and compound 51 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, 6-[(1-acetylazetidin-3-yl)amino]-N-[(2R)-2-hydroxy-2-{(3S)-7-[(4-methyl-1,3-oxazol-5-yl)methoxy]-1,2,3,4-tetrahydroisoquinolin-3-yl}ethyl]-2-(4-methylpiperidin-1-yl)pyrimidine-4-carboxamide, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2024-12-11 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | MTA-Cooperative PRMT5 Inhibitors: Mechanism Switching Through Structure-Based Design.
J.Med.Chem., 68, 2025
|
|
6MZC
 
 | | Human TFIID BC core | | Descriptor: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 2, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
7N0A
 
 | | Structure of Human Leukaemia Inhibitory Factor with Fab MSC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Leukemia inhibitory factor, ... | | Authors: | Raman, S, Bosch, A, Fransson, J, Julien, J.P. | | Deposit date: | 2021-05-25 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Therapeutic Targeting of LIF Overcomes Macrophage-mediated Immunosuppression of the Local Tumor Microenvironment.
Clin.Cancer Res., 29, 2023
|
|
9O4B
 
 | | Cryo-EM structure of in-vitro alpha-synuclein fibril bound with Exemplar-6 PET-radioligand | | Descriptor: | 2-(3,4-dimethylphenoxy)-N-[3-(4-fluorophenyl)-1,2-oxazol-5-yl]acetamide, Alpha-synuclein | | Authors: | Sanchez, J.C, Perez, R.M, Borcik, C.G, Guarino, D.S, Dhavala, D.D, Baumgardt, J.K, Shaffer, K.D, Kotzbauer, P.T, Mach, R.H, Rienstra, C.M, Petersson, E.J, Wright, E.R. | | Deposit date: | 2025-04-08 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Complementary Structural and Chemical Biology Methods Reveal the Basis for Selective Radioligand Binding to Alpha-Synuclein in MSA Tissue
To Be Published
|
|
7AD8
 
 | | Core TFIIH-XPA-DNA complex with modelled p62 subunit | | Descriptor: | DNA (49-MER), DNA repair protein complementing XP-A cells, General transcription and DNA repair factor IIH helicase subunit XPB, ... | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2020-09-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The TFIIH subunits p44/p62 act as a damage sensor during nucleotide excision repair.
Nucleic Acids Res., 48, 2020
|
|
9GMR
 
 | | SIRT7-H3K36MTUnucleosome complex | | Descriptor: | DNA (149-MER), Histone H2A type 2-A, Histone H2B type 1-J, ... | | Authors: | Moreno-Yruela, C, Ekundayo, B, Foteva, P, Calvino-Sanles, E, Ni, D, Stahlberg, H, Fierz, B. | | Deposit date: | 2024-08-29 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of SIRT7 nucleosome engagement and substrate specificity.
Nat Commun, 16, 2025
|
|
8H3Q
 
 | | Cryo-EM Structure of the CAND1-Cul3-Rbx1 complex | | Descriptor: | Cullin-3, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5N4W
 
 | | Crystal structure of the Cul2-Rbx1-EloBC-VHL ubiquitin ligase complex | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | Authors: | Cardote, T.A.F, Gadd, M.S, Ciulli, A. | | Deposit date: | 2017-02-11 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal Structure of the Cul2-Rbx1-EloBC-VHL Ubiquitin Ligase Complex.
Structure, 25, 2017
|
|
8I7R
 
 | | In situ structure of axonemal doublet microtubules in mouse sperm with 48-nm repeat | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Zhu, Y, Yin, G.L, Tai, L.H, Sun, F. | | Deposit date: | 2023-02-02 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | In-cell structural insight into the stability of sperm microtubule doublet.
Cell Discov, 9, 2023
|
|
1TX4
 
 | | RHO/RHOGAP/GDP(DOT)ALF4 COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, P50-RHOGAP, ... | | Authors: | Rittinger, K, Walker, P.A, Smerdon, S.J, Gamblin, S.J. | | Deposit date: | 1997-07-29 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue.
Nature, 389, 1997
|
|
8CDK
 
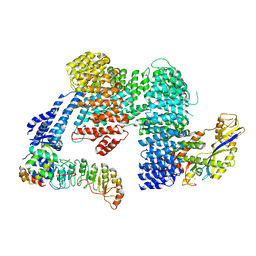 | |
8CDJ
 
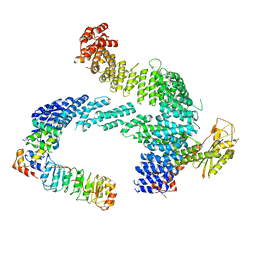 | | CAND1 b-hairpin++-SCF-SKP2 CAND1 rolling SCF engaged | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2023-01-31 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
5LPX
 
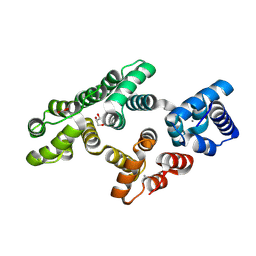 | | Crystal structure of PKC phosphorylation-mimicking mutant (S26E) Annexin A2 | | Descriptor: | Annexin A2, CALCIUM ION, GLYCEROL | | Authors: | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
5LQ0
 
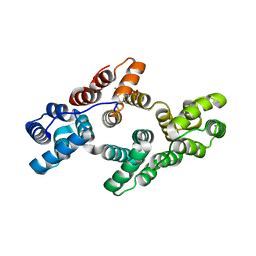 | | Crystal structure of Tyr24 phosphorylated Annexin A2 at 2.9 A resolution | | Descriptor: | Annexin A2, CALCIUM ION | | Authors: | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
8H36
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H37
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a tetrameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.52 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5LPU
 
 | | Crystal structure of Annexin A2 complexed with S100A4 | | Descriptor: | Annexin A2, CALCIUM ION, GLYCEROL, ... | | Authors: | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
5LQ2
 
 | | Crystal structure of Tyr24 phosphorylated Annexin A2 at 3.4 A resolution | | Descriptor: | Annexin A2, CALCIUM ION | | Authors: | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
