1E5G
 
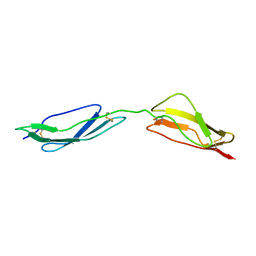 | | Solution structure of central CP module pair of a pox virus complement inhibitor | | Descriptor: | COMPLEMENT CONTROL PROTEIN C3 | | Authors: | Henderson, C.E, Bromek, K, Mullin, N.P, Smith, B.O, Uhrin, D, Barlow, P.N. | | Deposit date: | 2000-07-25 | | Release date: | 2000-08-31 | | Last modified: | 2013-07-03 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Central Ccp Module Pair of a Poxvirus Complement Control Protein
J.Mol.Biol., 307, 2001
|
|
1E5H
 
 | | DELTA-R307A DEACETOXYCEPHALOSPORIN C SYNTHASE COMPLEXED WITH SUCCINATE AND CARBON DIOXIDE | | Descriptor: | CARBON DIOXIDE, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (II) ION, ... | | Authors: | Lee, H.J, Lloyd, M.D, Harlos, K, Clifton, I.J, Baldwin, J.E, Schofield, C.J. | | Deposit date: | 2000-07-26 | | Release date: | 2001-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Kinetic and Crystallographic Studies on Deacetoxycephalosporin C Synthase (Daocs)
J.Mol.Biol., 308, 2001
|
|
1E5I
 
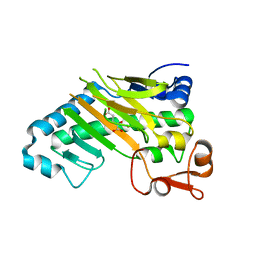 | | DELTA-R306 DEACETOXYCEPHALOSPORIN C SYNTHASE COMPLEXED WITH IRON AND 2-OXOGLUTARATE. | | Descriptor: | 2-OXOGLUTARIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (II) ION | | Authors: | Lee, H.J, Lloyd, M.D, Harlos, K, Clifton, I.J, Baldwin, J.E, Schofield, C.J. | | Deposit date: | 2000-07-26 | | Release date: | 2001-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and Crystallographic Studies on Deacetoxycephalosporin C Synthase (Daocs)
J.Mol.Biol., 308, 2001
|
|
1E5J
 
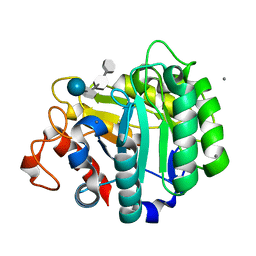 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE TETRAGONAL CRYSTAL FORM IN COMPLEX WITH METHYL-4II-S-ALPHA-CELLOBIOSYL-4II-THIO-BETA-CELLOBIOSIDE | | Descriptor: | CALCIUM ION, ENDOGLUCANASE 5A, alpha-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside | | Authors: | Fort, S, Varrot, A, Schulein, M, Cottaz, S, Driguez, H, Davies, G.J. | | Deposit date: | 2000-07-26 | | Release date: | 2001-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mixed-Linkage Cellooligosaccharides: A New Class of Glycoside Hydrolase Inhibitors
Chembiochem, 2, 2001
|
|
1E5K
 
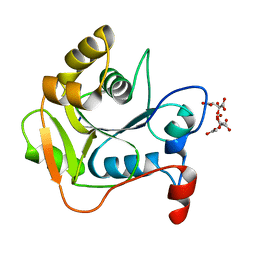 | | CRYSTAL STRUCTURE OF THE MOLYBDENUM COFACTOR BIOSYNTHESIS PROTEIN MOBA (PROTEIN FA) FROM ESCHERICHIA COLI AT NEAR ATOMIC RESOLUTION | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Stevenson, C.E.M, Sargent, F, Buchanan, G, Palmer, T, Lawson, D.M. | | Deposit date: | 2000-07-27 | | Release date: | 2000-11-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of the Molybdenum Cofactor Biosynthesis Protein Moba from Escherichia Coli at Near Atomic Resolution
Structure, 8, 2000
|
|
1E5L
 
 | | Apo saccharopine reductase from Magnaporthe grisea | | Descriptor: | SACCHAROPINE REDUCTASE | | Authors: | Johansson, E, Steffens, J.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 2000-07-27 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Saccharopine Reductase from Magnaporthe Grisea, an Enzyme of the Alpha-Aminoadipate Pathway of Lysine Biosynthesis
Structure, 8, 2000
|
|
1E5M
 
 | |
1E5N
 
 | | E246C mutant of P fluorescens subsp. cellulosa xylanase A in complex with xylopentaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Lo Leggio, L, Jenkins, J.A, Harris, G.W, Pickersgill, R.W. | | Deposit date: | 2000-07-27 | | Release date: | 2000-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray crystallographic study of xylopentaose binding to Pseudomonas fluorescens xylanase A.
Proteins, 41, 2000
|
|
1E5O
 
 | | Endothiapepsin complex with inhibitor DB2 | | Descriptor: | ENDOTHIAPEPSIN, N-[(2S)-2-amino-3-phenylpropyl]-D-methionyl-L-alanyl-L-isoleucine | | Authors: | Read, J.A, Cooper, J.B, Toldo, L, Bailey, D. | | Deposit date: | 2000-07-28 | | Release date: | 2000-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Refinement of Four Endothiapepsin Inhibitor Complexes. Crystallographic Studies of Cytochrome Ch from Methylobacterium Extorquens and Inhibitor Complexes of Aspartic Proteinases.
Thesis, 1999
|
|
1E5P
 
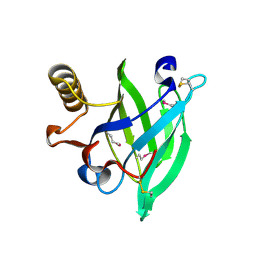 | | Crystal structure of aphrodisin, a sex pheromone from female hamster | | Descriptor: | APHRODISIN | | Authors: | Vincent, F, Brown, K, Spinelli, S, Cambillau, C, Tegoni, M. | | Deposit date: | 2000-07-28 | | Release date: | 2001-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of aphrodisin, a sex pheromone from female hamster.
J.Mol.Biol., 305, 2001
|
|
1E5Q
 
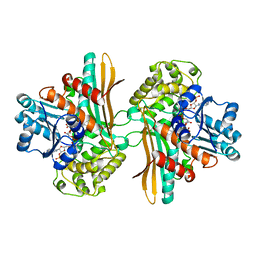 | | Ternary complex of saccharopine reductase from Magnaporthe grisea, NADPH and saccharopine | | Descriptor: | N-(5-AMINO-5-CARBOXYPENTYL)GLUTAMIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Saccharopine dehydrogenase [NADP(+), ... | | Authors: | Johansson, E, Steffens, J.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 2000-07-28 | | Release date: | 2000-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Saccharopine Reductase from Magnaporthe Grisea, an Enzyme of the Alpha-Aminoadipate Pathway of Lysine Biosynthesis
Structure, 8, 2000
|
|
1E5R
 
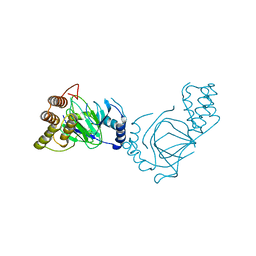 | | Proline 3-hydroxylase (type II) -apo form | | Descriptor: | PROLINE OXIDASE | | Authors: | Clifton, I.J, Hsueh, L.C, Baldwin, J.E, Schofield, C.J, Harlos, K. | | Deposit date: | 2000-07-28 | | Release date: | 2001-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of proline 3-hydroxylase. Evolution of the family of 2-oxoglutarate dependent oxygenases.
Eur.J.Biochem., 268, 2001
|
|
1E5S
 
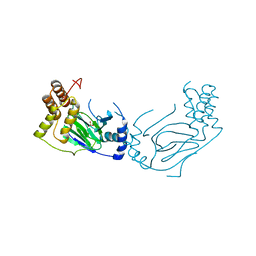 | | Proline 3-hydroxylase (type II) - Iron form | | Descriptor: | FE (II) ION, PROLINE OXIDASE, SULFATE ION | | Authors: | Clifton, I.J, Hsueh, L.C, Baldwin, J.E, Schofield, C.J, Harlos, K. | | Deposit date: | 2000-07-28 | | Release date: | 2001-07-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of proline 3-hydroxylase. Evolution of the family of 2-oxoglutarate dependent oxygenases.
Eur.J.Biochem., 268, 2001
|
|
1E5T
 
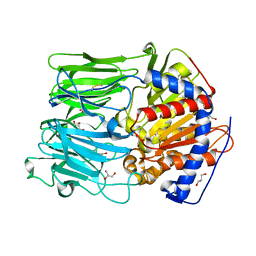 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, MUTANT | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Fulop, V. | | Deposit date: | 2000-08-02 | | Release date: | 2000-10-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalysis of Serine Oligopeptidases is Controlled by a Gating Filter Mechanism
Embo Rep., 1, 2000
|
|
1E5U
 
 | | NMR Representative Structure of Intimin-190 (Int190) from Enteropathogenic E. coli | | Descriptor: | INTIMIN | | Authors: | Prasannan, S, Matthews, S.J, Batchelor, M, Daniell, S, Reece, S, Frankel, G, Dougan, G, Connerton, I, Bloomberg, G. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-16 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Recognition of the Translocated Intimin Receptor (Tir) by Intimin from Enteropathogenic E. Coli
Embo J., 19, 2000
|
|
1E5V
 
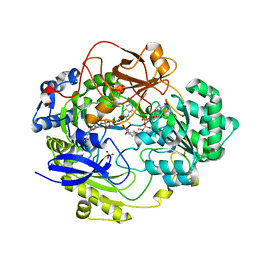 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
1E5W
 
 | |
1E5X
 
 | |
1E5Y
 
 | |
1E5Z
 
 | |
1E60
 
 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES - Structure II BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-06 | | Release date: | 2000-08-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
1E61
 
 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES - Structure II BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-06 | | Release date: | 2000-08-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
1E62
 
 | | Ferredoxin:NADP+ reductase mutant with Lys 75 replaced by Arg (K75R) | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2000-08-07 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Interactions for Complex Formation between Ferredoxin-Nadp+ Reductase and its Protein Partners.
Proteins, 59, 2005
|
|
1E63
 
 | | Ferredoxin:NADP+ Reductase Mutant with LYS 75 Replaced by SER (K75S) | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Gomez-Moreno, C. | | Deposit date: | 2000-08-07 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Interactions for Complex Formation between Ferredoxin-Nadp+ Reductase and its Protein Partners.
Proteins, 59, 2005
|
|
1E64
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LYS 75 REPLACED BY GLN (K75Q) | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2000-08-07 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Interactions for Complex Formation between Ferredoxin-Nadp+ Reductase and its Protein Partners.
Proteins, 59, 2005
|
|
