1EYL
 
 | | STRUCTURE OF A RECOMBINANT WINGED BEAN CHYMOTRYPSIN INHIBITOR | | Descriptor: | CHYMOTRYPSIN INHIBITOR, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Ghosh, S. | | Deposit date: | 2000-05-07 | | Release date: | 2000-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
Protein Eng., 14, 2001
|
|
4VGC
 
 | | GAMMA-CHYMOTRYPSIN D-NAPHTHYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | D-1-NAPHTHYL-2-ACETAMIDO-ETHANE BORONIC ACID, GAMMA CHYMOTRYPSIN, SULFATE ION | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
1FMZ
 
 | | CRYSTAL STRUCTURE OF A MUTANT WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN, N14K. | | Descriptor: | CHYMOTRYPSIN INHIBITOR 3, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Dasgupta, J, Ghosh, S. | | Deposit date: | 2000-08-19 | | Release date: | 2001-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
PROTEIN ENG., 14, 2001
|
|
1FN0
 
 | | STRUCTURE OF A MUTANT WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN, N14D. | | Descriptor: | CHYMOTRYPSIN INHIBITOR 3, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Dasgupta, J, Ghosh, S. | | Deposit date: | 2000-08-19 | | Release date: | 2001-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
PROTEIN ENG., 14, 2001
|
|
3SJH
 
 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-29) of drosophila Ciboulot and the C-terminal domain (residues 18-44) of bovine Thymosin-beta4, bound to G-actin-ATP-Latrunculin A | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-06-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Embo J., 31, 2012
|
|
5CHA
 
 | |
2VGC
 
 | | GAMMA-CHYMOTRYPSIN D-PARA-CHLORO-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | D-1-(4-CHLOROPHENYL)-2-(ACETAMIDO)ETHANE BORONIC ACID, GAMMA CHYMOTRYPSIN, SULFATE ION | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
5GCH
 
 | |
6CHA
 
 | |
7QIQ
 
 | | CRYSTAL STRUCTURE OF THE P1 aminobutanoic acid (ABU) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
7QIR
 
 | | CRYSTAL STRUCTURE OF THE P1 monofluorethylglycine(MfeGly) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
7QIS
 
 | | CRYSTAL STRUCTURE OF THE P1 difluoroethylglycine (DfeGly) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
7QIT
 
 | | CRYSTAL STRUCTURE OF THE P1 trifluoroethylglycine (TfeGly) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
1AB9
 
 | | CRYSTAL STRUCTURE OF BOVINE GAMMA-CHYMOTRYPSIN | | Descriptor: | GAMMA-CHYMOTRYPSIN, PENTAPEPTIDE (TPGVY), SULFATE ION | | Authors: | Sugio, S, Kashima, A, Inoue, Y, Maeda, I, Nose, T, Shimohigashi, Y. | | Deposit date: | 1997-02-05 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structure of a dipeptide-chymotrypsin complex in an inhibitory interaction.
Eur.J.Biochem., 255, 1998
|
|
6QPK
 
 | | Zt-KP6-1: an effector from Zymoseptoria tritici | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, SULFATE ION, ... | | Authors: | Padilla, A, Hoh, F, De guillen, K. | | Deposit date: | 2019-02-14 | | Release date: | 2020-03-04 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Zymoseptoria tritici effectors structurally related to killer proteins UmV-KP4 and UmV-KP6 are toxic to fungi, and define extended protein families in fungi
Biorxiv, 2024
|
|
8GTY
 
 | |
5GUS
 
 | | Crystal structure of ASCH domain from Zymomonas mobilis | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, Helix-turn-helix domain-containing protein, ... | | Authors: | Ha, S.C, Park, S.Y, Kim, J.S. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystal structure of an ASCH protein from Zymomonas mobilis and its ribonuclease activity specific for single-stranded RNA.
Sci Rep, 7, 2017
|
|
5GUQ
 
 | | Crystal structure of ASCH from Zymomonas mobilis | | Descriptor: | Helix-turn-helix domain-containing protein | | Authors: | Ha, S.C, Park, S.Y, Kim, J.S. | | Deposit date: | 2016-08-30 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Crystal structure of an ASCH protein from Zymomonas mobilis and its ribonuclease activity specific for single-stranded RNA.
Sci Rep, 7, 2017
|
|
1ACB
 
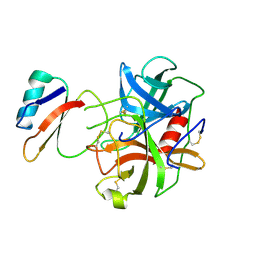 | | CRYSTAL AND MOLECULAR STRUCTURE OF THE BOVINE ALPHA-CHYMOTRYPSIN-EGLIN C COMPLEX AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | ALPHA-CHYMOTRYPSIN, Eglin C | | Authors: | Bolognesi, M, Frigerio, F, Coda, A, Pugliese, L, Lionetti, C, Menegatti, E, Amiconi, G, Schnebli, H.P, Ascenzi, P. | | Deposit date: | 1991-11-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal and molecular structure of the bovine alpha-chymotrypsin-eglin c complex at 2.0 A resolution.
J.Mol.Biol., 225, 1992
|
|
5EUJ
 
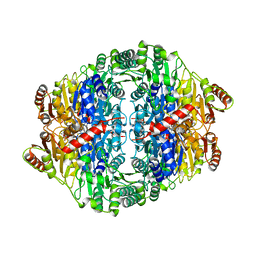 | | PYRUVATE DECARBOXYLASE FROM ZYMOBACTER PALMAE | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Pyruvate decarboxylase, ... | | Authors: | Buddrus, L, Crennell, S.J, Leak, D.J, Danson, M.J, Andrews, E.S.V, Arcus, V.L. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of pyruvate decarboxylase from Zymobacter palmae.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2GCT
 
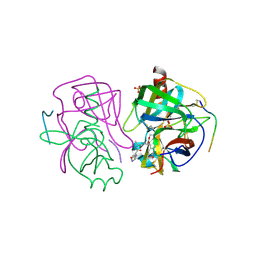 | |
7KU0
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 138 (yellow) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU2
 
 | | Data clustering and dynamics of chymotrypsinogen clulster 140 (structure) | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU1
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 139 (green) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU3
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 141 (cyan) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
