7EPW
 
 | | Crystal structure of monooxygenase Tet(X4) with tigecycline | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase, TIGECYCLINE | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2021-04-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and mechanistic basis of the high catalytic activity of monooxygenase Tet(X4) on tigecycline.
Bmc Biol., 19, 2021
|
|
7EPV
 
 | | Crystal structure of tigecycline degrading monooxygenase Tet(X4) | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase, GLYCEROL | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2021-04-27 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and mechanistic basis of the high catalytic activity of monooxygenase Tet(X4) on tigecycline.
Bmc Biol., 19, 2021
|
|
4K22
 
 | | Structure of the C-terminal truncated form of E.Coli C5-hydroxylase UBII involved in ubiquinone (Q8) biosynthesis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pecqueur, L, Lombard, M, Golinelli-pimpaneau, B, Fontecave, M. | | Deposit date: | 2013-04-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ubiI, a New Gene in Escherichia coli Coenzyme Q Biosynthesis, Is Involved in Aerobic C5-hydroxylation.
J.Biol.Chem., 288, 2013
|
|
7FCO
 
 | | ChlB4 Halogenase | | Descriptor: | CHLORIDE ION, ChlB4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Saeed, A.U, Zheng, J. | | Deposit date: | 2021-07-15 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal insight of FAD-dependent bifunctional halogenase ChlB4 in the biosynthesis of Chlorothricin
To Be Published
|
|
2C4C
 
 | | Crystal structure of the NADPH-treated monooxygenase domain of MICAL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NEDD9-INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS | | Authors: | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2005-10-18 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BRY
 
 | | Crystal structure of the native monooxygenase domain of MICAL at 1.45 A resolution | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2005-05-13 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3RP6
 
 | | Crystal Structure of Klebsiella pneumoniae HpxO complexed with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, flavoprotein monooxygenase | | Authors: | Hicks, K.A, O'Leary, S.E, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-16 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Studies of HpxO, a Novel Flavin Adenine Dinucleotide-Dependent Urate Oxidase from Klebsiella pneumoniae.
Biochemistry, 52, 2013
|
|
3RP7
 
 | | Crystal Structure of Klebsiella pneumoniae HpxO complexed with FAD and uric acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, URIC ACID, flavoprotein monooxygenase | | Authors: | Hicks, K.A, O'Leary, S.E, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-16 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structural and Mechanistic Studies of HpxO, a Novel Flavin Adenine Dinucleotide-Dependent Urate Oxidase from Klebsiella pneumoniae.
Biochemistry, 52, 2013
|
|
8GSM
 
 | | Crystal Structure of VibMO1 | | Descriptor: | 4-hydroxybenzoate decarboxylase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Ying, Z, Feng, K.N. | | Deposit date: | 2022-09-06 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal Structure of VibMO1
To Be Published
|
|
3RP8
 
 | | Crystal Structure of Klebsiella pneumoniae R204Q HpxO complexed with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, flavoprotein monooxygenase | | Authors: | Hicks, K.A, O'Leary, S.E, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Structural and Mechanistic Studies of HpxO, a Novel Flavin Adenine Dinucleotide-Dependent Urate Oxidase from Klebsiella pneumoniae.
Biochemistry, 52, 2013
|
|
1K0L
 
 | | Pseudomonas aeruginosa phbh R220Q free of p-OHB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, SULFATE ION, ... | | Authors: | Wang, J, Ortiz-Maldonado, M, Entsch, B, Ballou, D, Gatti, D.L. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IUU
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE AT PH 9.4 | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
1IUS
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE AT PH 5.0 | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
1IUV
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-4-HYDROXYBENZOATE AT PH 5.0 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
1IUX
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-4-HYDROXYBENZOATE AT PH 9.4 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
1IUT
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE AT PH 7.4 | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
5EOW
 
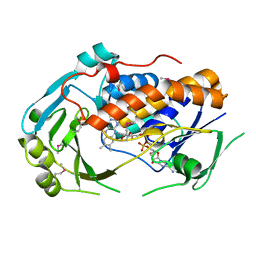 | | Crystal Structure of 6-Hydroxynicotinic Acid 3-Monooxygenase from Pseudomonas putida KT2440 | | Descriptor: | 6-hydroxynicotinate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yuen, M.E, Zhen, W, Gerwig, T.J, Story, R.W, Kopp, M, Nakamoto, K, Snider, M.J, Hicks, K.A. | | Deposit date: | 2015-11-10 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Characterization of 6-Hydroxynicotinic Acid 3-Monooxygenase, A Novel Decarboxylative Hydroxylase Involved in Aerobic Nicotinate Degradation.
Biochemistry, 55, 2016
|
|
1IUW
 
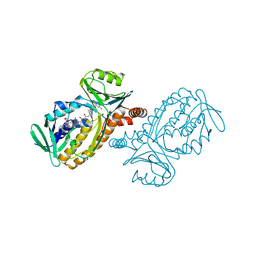 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-4-HYDROXYBENZOATE AT PH 7.4 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
5EVY
 
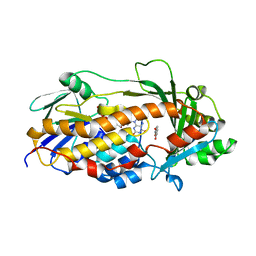 | | Salicylate hydroxylase substrate complex | | Descriptor: | 2-HYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Salicylate hydroxylase | | Authors: | Morimoto, Y, Uemura, T. | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The catalytic mechanism of decarboxylative hydroxylation of salicylate hydroxylase revealed by crystal structure analysis at 2.5 angstrom resolution
Biochem.Biophys.Res.Commun., 469, 2016
|
|
2X3N
 
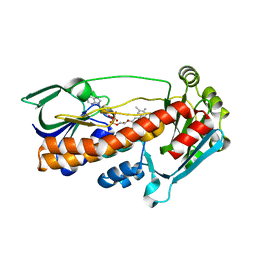 | | Crystal structure of pqsL, a probable FAD-dependent monooxygenase from Pseudomonas aeruginosa | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROBABLE FAD-DEPENDENT MONOOXYGENASE | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
5FN0
 
 | | Crystal structure of Pseudomonas fluorescens kynurenine-3- monooxygenase (KMO) in complex with GSK180 | | Descriptor: | 3-(5,6-DICHLORO-2-OXOBENZO[D]OXAZOL-3(2H)-YL)PROPANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, KYNURENINE 3-MONOOXYGENASE | | Authors: | Mole, D.J, Webster, S.P, Uings, I, Zheng, X, Binnie, M, Wilson, K, Hutchinson, J.P, Mirguet, O, Walker, A, Beaufils, B, Ancellin, N, Trottet, L, Beneton, V, Mowat, C.G, Wilkinson, M, Rowland, P, Haslam, C, McBride, A, Homer, N.Z.M, Baily, J.E, Sharp, M.G.F, Garden, O.J, Hughes, J, Howie, S.E.M, Holmes, D, Liddle, J, Iredale, J.P. | | Deposit date: | 2015-11-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Kynurenine-3-Monooxygenase Inhibition Prevents Multiple Organ Failure in Rodent Models of Acute Pancreatitis.
Nat.Med. (N.Y.), 22, 2016
|
|
7ON9
 
 | | Crystal structure of para-hydroxybenzoate-3-hydroxylase PraI | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Zahn, M, McGeehan, J.E. | | Deposit date: | 2021-05-25 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Debottlenecking 4-hydroxybenzoate hydroxylation in Pseudomonas putida KT2440 improves muconate productivity from p-coumarate.
Metab Eng, 70, 2022
|
|
8TWG
 
 | |
8TWF
 
 | |
8UIQ
 
 | | H47Q NicC with 2-mercaptopyridine ligand | | Descriptor: | 2-PYRIDINETHIOL, 6-hydroxynicotinate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hicks, K.A, Perry, K, Turlington, Z.R, Vaz Ferreira de Macedo, S. | | Deposit date: | 2023-10-10 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Ligand bound structure of a 6-hydroxynicotinic acid 3-monooxygenase provides mechanistic insights.
Arch.Biochem.Biophys., 752, 2024
|
|
