5MXP
 
 | |
5O2G
 
 | | Crystal structure determination from picosecond infrared laser ablated protein crystals by serial synchrotron crystallography | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Kaub, J, Busse, F, Mehrabi, P, Mueller-Werkmeiser, H, Pai, E.F, Robertson, W.D, Miller, R.J.D. | | Deposit date: | 2017-05-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure determination from picosecond infrared laser ablated protein crystals by serial synchrotron crystallography
To Be Published
|
|
5O2I
 
 | | An efficient setup for fixed-target, time-resolved serial crystallography with optical excitation | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Mueller-Werkmeister, H, Mehrabi, P, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2017-05-20 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An efficient setup for fixed-target, time-resolved serial crystallography with optical excitation
To Be Published
|
|
7NFZ
 
 | |
5NYV
 
 | | Crystal structure determination from picosecond infrared laser ablated protein crystals by serial synchrotron crystallography | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Kaub, J, Busse, F, Mehrabi, P, Mueller-Werkmeiser, H, Pai, E.F, Robertson, W.D, Miller, R.J.D. | | Deposit date: | 2017-05-11 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein crystals IR laser ablated from aqueous solution at high speed retain their diffractive properties: applications in high-speed serial crystallography.
J.Appl.Crystallogr., 50, 2017
|
|
1IZ8
 
 | | Re-refinement of the structure of hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis UT26 with 1,3-propanediol, a product of debromidation of dibrompropane, at 2.0A resolution | | Descriptor: | 1,3-PROPANDIOL, BROMIDE ION, CALCIUM ION, ... | | Authors: | Streltsov, V.A. | | Deposit date: | 2002-09-30 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates
Biochemistry, 42, 2003
|
|
1IUN
 
 | | meta-Cleavage product hydrolase from Pseudomonas fluorescens IP01 (CumD) S103A mutant hexagonal | | Descriptor: | ACETATE ION, meta-Cleavage product hydrolase | | Authors: | Fushinobu, S, Saku, T, Hidaka, M, Jun, S.-Y, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a meta-cleavage product hydrolase from Pseudomonas
fluorescens IP01 (CumD) complexed with cleavage products
PROTEIN SCI., 11, 2002
|
|
8HGW
 
 | | Crystal structure of MehpH in complex with MBP | | Descriptor: | 1-BUTANOL, Monoalkyl phthalate hydrolase, PHTHALIC ACID | | Authors: | Zhang, Z.M, Wang, Y.J, Chen, Y.B. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.80001163 Å) | | Cite: | Molecular insights into the catalytic mechanism of plasticizer degradation by a monoalkyl phthalate hydrolase.
Commun Chem, 6, 2023
|
|
8HGV
 
 | |
1J1I
 
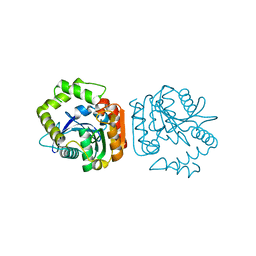 | | Crystal structure of a His-tagged Serine Hydrolase Involved in the Carbazole Degradation (CarC enzyme) | | Descriptor: | meta cleavage compound hydrolase | | Authors: | Habe, H, Morii, K, Fushinobu, S, Nam, J.W, Ayabe, Y, Yoshida, T, Wakagi, T, Yamane, H, Nojiri, H, Omori, T. | | Deposit date: | 2002-12-05 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of a histidine-tagged serine hydrolase involved in the carbazole degradation (CarC enzyme).
Biochem.Biophys.Res.Commun., 303, 2003
|
|
8IK2
 
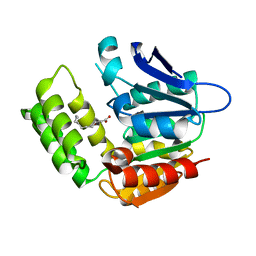 | | RhlA exhibits dual thioesterase and acyltransferase activities during rhamnolipid biosynthesis | | Descriptor: | (3~{S})-3-oxidanyldecanoic acid, 3-(3-hydroxydecanoyloxy)decanoate synthase | | Authors: | Tang, T, Fu, L.H, Xie, W.H, Luo, Y.Z, Zhang, Y.T, Si, T. | | Deposit date: | 2023-02-28 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | RhlA Exhibits Dual Thioesterase and Acyltransferase Activities during Rhamnolipid Biosynthesis
Acs Catalysis, 13, 2023
|
|
1IUO
 
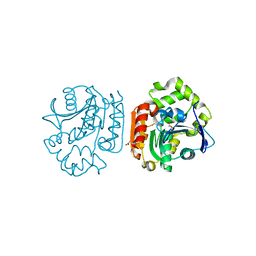 | | meta-Cleavage product hydrolase from Pseudomonas fluorescens IP01 (CumD) S103A mutant complexed with acetates | | Descriptor: | ACETATE ION, meta-Cleavage product hydrolase | | Authors: | Fushinobu, S, Saku, T, Hidaka, M, Jun, S.-Y, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of a meta-cleavage product hydrolase from Pseudomonas
fluorescens IP01 (CumD) complexed with cleavage products
PROTEIN SCI., 11, 2002
|
|
7F5W
 
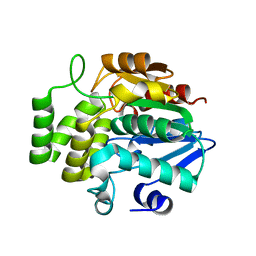 | |
8HGU
 
 | |
8HM5
 
 | |
8OE2
 
 | | Structure of hyperstable haloalkane dehalogenase variant DhaA223 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marek, M. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Advancing Enzyme's Stability and Catalytic Efficiency through Synergy of Force-Field Calculations, Evolutionary Analysis, and Machine Learning.
Acs Catalysis, 13, 2023
|
|
6F9O
 
 | | Crystal structure of cold-adapted haloalkane dehalogenase DpcA from Psychrobacter cryohalolentis K5 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Haloalkane dehalogenase, ... | | Authors: | Tratsiak, K, Prudnikova, T, Drienovska, I, Damborsky, J, Brynda, J, Pachl, P, Kuty, M, Chaloupkova, R, Kuta Smatanova, I, Rezacova, P. | | Deposit date: | 2017-12-15 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure of the cold-adapted haloalkane dehalogenase DpcA from Psychrobacter cryohalolentis K5.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8JLV
 
 | |
8G48
 
 | |
8G49
 
 | | FphE, Staphylococcus aureus fluorophosphonate-binding serine hydrolases E, Oxadiazolone compound 3 bound | | Descriptor: | Fluorophosphonate-binding serine hydrolase E, methyl 2-formyl-2-[3-methyl-4-(3-phenoxybenzamido)phenyl]hydrazine-1-carboxylate | | Authors: | Fellner, M, Bakker, A.T, Martin, N.I, Stelt, M. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | FphE, Staphylococcus aureus fluorophosphonate-binding serine hydrolases E, Oxadiazolone compound 3 bound
To be published
|
|
6FSX
 
 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-werkmeiser, H, Tellkamp, F, Jha, A, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E, Miller, D. | | Deposit date: | 2018-02-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
6FR2
 
 | | Soluble epoxide hydrolase in complex with LK864 | | Descriptor: | 1-[(4~{S})-9-propan-2-ylsulfonyl-1-oxa-9-azaspiro[5.5]undecan-4-yl]-3-[[4-(trifluoromethyloxy)phenyl]methyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | Authors: | Kramer, J.S, Pogoryelov, D, Krasavin, M, Proschak, E. | | Deposit date: | 2018-02-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Discovery of polar spirocyclic orally bioavailable urea inhibitors of soluble epoxide hydrolase.
Bioorg. Chem., 80, 2018
|
|
8JY1
 
 | |
7JQZ
 
 | |
7JQX
 
 | |
