2FIW
 
 | | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris | | Descriptor: | ACETYL COENZYME *A, GCN5-related N-acetyltransferase:Aminotransferase, class-II, ... | | Authors: | Kim, Y, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-30 | | Release date: | 2006-02-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris
To be Published, 2006
|
|
1PU9
 
 | | Crystal Structure of Tetrahymena GCN5 with Bound Coenzyme A and a 19-residue Histone H3 Peptide | | Descriptor: | COENZYME A, HAT A1, Histone H3 | | Authors: | Clements, A, Poux, A.N, Lo, W.S, Pillus, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for histone and phospho-histone binding by the GCN5 histone acetyltransferase
Mol.Cell, 12, 2003
|
|
1PUA
 
 | | Crystal Structure of Tetrahymena GCN5 with Bound Coenzyme A and a Phosphorylated, 19-residue Histone H3 peptide | | Descriptor: | COENZYME A, HAT A1, Histone H3 | | Authors: | Clements, A, Poux, A.N, Lo, W.S, Pillus, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for histone and phospho-histone binding by the GCN5 histone acetyltransferase
Mol.Cell, 12, 2003
|
|
2Q0Y
 
 | |
2OZG
 
 | |
3TT2
 
 | | Crystal Structure of GCN5-related N-Acetyltransferase from Sphaerobacter thermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GCN5-related N-acetyltransferase, GLYCEROL, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structure of GCN5-related N-Acetyltransferase from Sphaerobacter thermophilus
To be Published
|
|
1R57
 
 | | NMR Solution Structure of a GCN5-like putative N-acetyltransferase from Staphylococcus aureus. Northeast Structural Genomics Consortium Target ZR31 | | Descriptor: | conserved hypothetical protein | | Authors: | Cort, J.R, Acton, T.B, Ma, L, Xiao, R.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-10-09 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an acetyl-CoA binding protein from Staphylococcus aureus representing a novel subfamily of GCN5-related N-acetyltransferase-like proteins.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
8XUW
 
 | |
4MY3
 
 | | Crystal Structure of GCN5-related N-acetyltransferase from Kribbella flavida | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GCN5-related N-acetyltransferase | | Authors: | Kim, Y, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-27 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure of GCN5-related N-acetyltransferase from Kribbella flavida
To be Published
|
|
4MY0
 
 | | Crystal Structure of GCN5-related N-acetyltransferase from Kribbella flavida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYL COENZYME *A, GCN5-related N-acetyltransferase, ... | | Authors: | Kim, Y, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal Structure of GCN5-related N-acetyltransferase from Kribbella flavida
To be Published
|
|
7KPP
 
 | | Structure of the E102A mutant of a GNAT superfamily PA3944 acetyltransferase | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase PA3944, COENZYME A, ... | | Authors: | Czub, M.P, Porebski, P.J, Majorek, K.A, Cymborowski, M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
7KPS
 
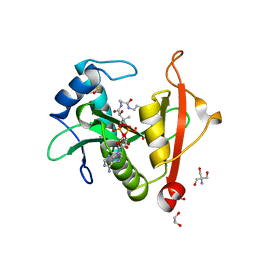 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with AcCoA | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYL COENZYME *A, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Reidl, C.T, Becker, D.P, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
5VDB
 
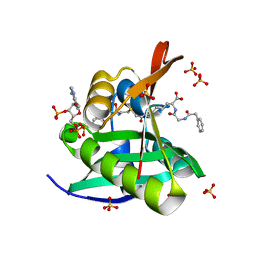 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with bisubstrate analog 3 | | Descriptor: | (3R,5S,9R,26S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14,20-trioxo-26-({[(phenylacetyl)amino]acetyl}amino)-2,4,6-trioxa-18-thia-11,15,21-triaza-3,5-diphosphaheptacosan-27-oic acid 3,5-dioxide (non-preferred name), SULFATE ION, acetyltransferase PA4794 | | Authors: | Majorek, K.A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-04-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Generating enzyme and radical-mediated bisubstrates as tools for investigating Gcn5-related N-acetyltransferases.
FEBS Lett., 591, 2017
|
|
5VD6
 
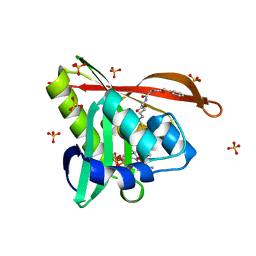 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with bisubstrate analog 6 | | Descriptor: | (3R,5S,9R,23S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14-dioxo-23-({[(phenylacetyl)amino]acetyl}amino)-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatetracosan-24-oic acid 3,5-dioxide (non-preferred name), SULFATE ION, acetyltransferase PA4794 | | Authors: | Majorek, K.A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-04-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Generating enzyme and radical-mediated bisubstrates as tools for investigating Gcn5-related N-acetyltransferases.
FEBS Lett., 591, 2017
|
|
8E6O
 
 | | Crystal structure of human GCN5 histone acetyltransferase domain | | Descriptor: | Histone acetyltransferase KAT2A, S-{(3S,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5,10,14-tetraoxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (2R)-2-hydroxypropanethioate | | Authors: | Lu, X.T, Tao, Y.J. | | Deposit date: | 2022-08-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of human GCN5 histone acetyltransferase domain
To Be Published
|
|
1YK3
 
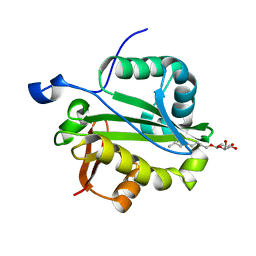 | | Crystal structure of Rv1347c from Mycobacterium tuberculosis | | Descriptor: | Hypothetical protein Rv1347c/MT1389, octyl beta-D-glucopyranoside | | Authors: | Card, G.L, Peterson, N.A, Smith, C.A, Rupp, B, Schick, B.M, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-01-16 | | Release date: | 2005-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Rv1347c, a putative antibiotic resistance protein from Mycobacterium tuberculosis, reveals a GCN5-related fold and suggests an alternative function in siderophore biosynthesis
J.Biol.Chem., 280, 2005
|
|
6EDV
 
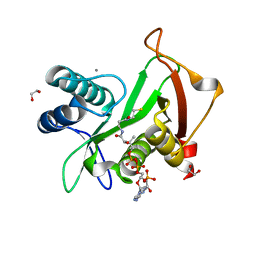 | | Structure of a GNAT superfamily acetyltransferase PA3944 in complex with CoA | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase PA3944, CALCIUM ION, ... | | Authors: | Majorek, K.A, Satchell, K.J.F, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-12 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Gcn5-Related N-Acetyltransferase (GNAT) Capable of Acetylating Polymyxin B and Colistin Antibiotics in Vitro.
Biochemistry, 57, 2018
|
|
1Z4R
 
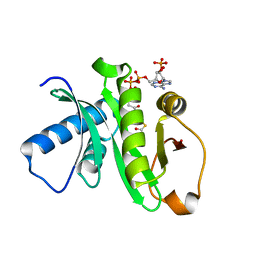 | | Human GCN5 Acetyltransferase | | Descriptor: | ACETYL COENZYME *A, General control of amino acid synthesis protein 5-like 2 | | Authors: | Dong, A, Bernstein, G, Schuetz, A, Antoshenko, T, Wu, H, Loppnau, P, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Plotnikov, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-16 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of a binary complex between human GCN5 histone acetyltransferase domain and acetyl coenzyme A
Proteins, 68, 2007
|
|
5XXS
 
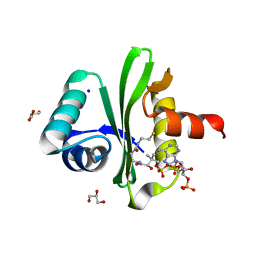 | |
5XXR
 
 | |
3PGP
 
 | | Crystal structure of PA4794 - GNAT superfamily protein in complex with AcCoA | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa.
J.Biol.Chem., 288, 2013
|
|
6EDD
 
 | | Crystal structure of a GNAT Superfamily PA3944 acetyltransferase in complex with CoA (P1 space group) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetyltransferase PA3944, ... | | Authors: | Czub, M.P, Porebski, P.J, Majorek, K.A, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-09 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Gcn5-Related N-Acetyltransferase (GNAT) Capable of Acetylating Polymyxin B and Colistin Antibiotics in Vitro.
Biochemistry, 57, 2018
|
|
5TPX
 
 | | Bromodomain from Plasmodium Faciparum Gcn5, complexed with compound | | Descriptor: | (1S,2S)-N~1~,N~1~-dimethyl-N~2~-(3-methyl[1,2,4]triazolo[3,4-a]phthalazin-6-yl)-1-phenylpropane-1,2-diamine, CHLORIDE ION, Histone acetyltransferase GCN5, ... | | Authors: | Lin, Y.H, Hou, C.F.D, MOUSTAKIM, M, DIXON, D.J, Loppnau, P, Tempel, W, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, BRENNAN, P.E, Walker, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a PCAF Bromodomain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MKX
 
 | | 1.68A STRUCTURE PCAF BROMODOMAIN WITH 4-chloro-2-methyl-5-(methylamino)pyridazin-3(2H)-one | | Descriptor: | 1,4-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-chloranyl-2-methyl-5-(methylamino)pyridazin-3-one, ... | | Authors: | Chung, C.-W. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of a Potent, Cell Penetrant, and Selective p300/CBP-Associated Factor (PCAF)/General Control Nonderepressible 5 (GCN5) Bromodomain Chemical Probe.
J. Med. Chem., 60, 2017
|
|
5MKY
 
 | | BROMODOMAIN OF HUMAN BRD9 WITH 4-chloro-2-methyl-5-((2-methyl-1,2,3,4-tetrahydroisoquinolin-5-yl)amino)pyridazin-3(2H)-one | | Descriptor: | 4-chloranyl-2-methyl-5-[(2-methyl-3,4-dihydro-1~{H}-isoquinolin-5-yl)amino]pyridazin-3-one, Bromodomain-containing protein 9 | | Authors: | Chung, C.-W. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of a Potent, Cell Penetrant, and Selective p300/CBP-Associated Factor (PCAF)/General Control Nonderepressible 5 (GCN5) Bromodomain Chemical Probe.
J. Med. Chem., 60, 2017
|
|
