6ZQD
 
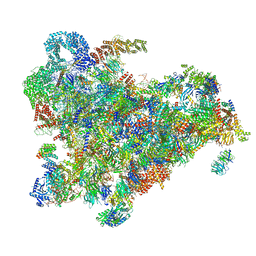 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Post-A1 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6QX9
 
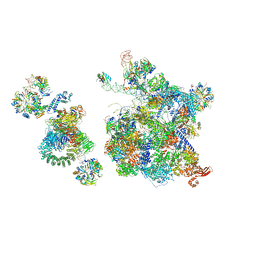 | | Structure of a human fully-assembled precatalytic spliceosome (pre-B complex). | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, AdML pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Charenton, C, Wilkinson, M.E, Nagai, K. | | Deposit date: | 2019-03-07 | | Release date: | 2019-04-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Mechanism of 5' splice site transfer for human spliceosome activation.
Science, 364, 2019
|
|
6ND4
 
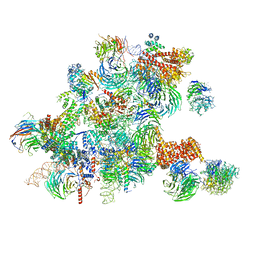 | | Conformational switches control early maturation of the eukaryotic small ribosomal subunit | | Descriptor: | 18S rRNA 5' domain start, 5'ETS rRNA, Bud21, ... | | Authors: | Hunziker, M, Barandun, J, Klinge, S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Conformational switches control early maturation of the eukaryotic small ribosomal subunit.
Elife, 8, 2019
|
|
6ZQA
 
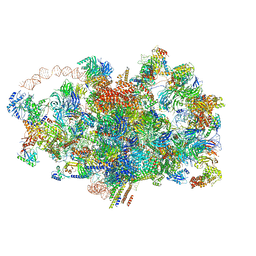 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state A (Poly-Ala) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6LQT
 
 | |
6ZQE
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-A (Poly-Ala) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
8FKV
 
 | |
8FKZ
 
 | |
8FKT
 
 | |
8FKX
 
 | |
8FKU
 
 | |
7YCX
 
 | | The structure of INTAC-PEC complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1,DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Zheng, H, Jin, Q, Wang, X, Qi, Y, Liu, W, Ren, Y, Zhao, D, Chen, F.X, Cheng, J, Chen, X, Xu, Y. | | Deposit date: | 2022-07-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural basis of INTAC-regulated transcription.
Protein Cell, 14, 2023
|
|
5MQF
 
 | | Cryo-EM structure of a human spliceosome activated for step 2 of splicing (C* complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ATP-dependent RNA helicase DHX8, Cell division cycle 5-like protein, ... | | Authors: | Bertram, K, Hartmuth, K, Kastner, B. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM structure of a human spliceosome activated for step 2 of splicing.
Nature, 542, 2017
|
|
9EP1
 
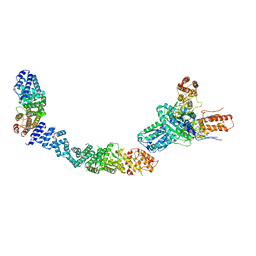 | |
9EP4
 
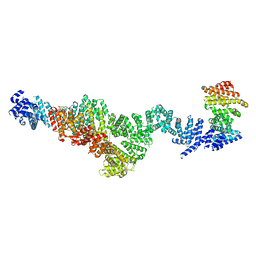 | | Structure of Integrator subcomplex INTS5/8/15 | | Descriptor: | Integrator complex subunit 15, Integrator complex subunit 5, Integrator complex subunit 8 | | Authors: | Razew, M, Galej, W.P. | | Deposit date: | 2024-03-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of the Integrator complex assembly and association with transcription factors.
Mol.Cell, 84, 2024
|
|
9FA4
 
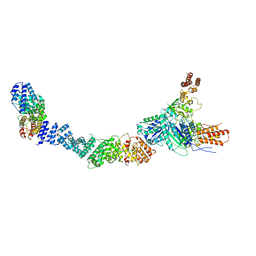 | |
8FL0
 
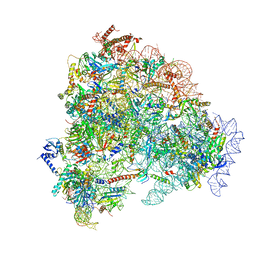 | |
9EOC
 
 | |
9FA7
 
 | |
9EOF
 
 | | Structure of the human INTS5/8/10/15 subcomplex | | Descriptor: | Integrator complex subunit 10, Integrator complex subunit 15, Integrator complex subunit 5, ... | | Authors: | Razew, M, Galej, W.P. | | Deposit date: | 2024-03-14 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural basis of the Integrator complex assembly and association with transcription factors.
Mol.Cell, 84, 2024
|
|
1JTF
 
 | | Crystal Structure Analysis of VP39-F180W mutant and m7GpppG complex | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Oguro, A, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2001-08-20 | | Release date: | 2002-07-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The "cap-binding slot" of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap.
Biochemistry, 41, 2002
|
|
1JSZ
 
 | | Crystal Structure Analysis of N7,9-dimethylguanine-VP39 complex | | Descriptor: | 7,9-DIMETHYLGUANINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Oguro, A, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2001-08-19 | | Release date: | 2002-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The "cap-binding slot" of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap.
Biochemistry, 41, 2002
|
|
1JTE
 
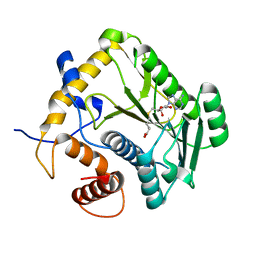 | | Crystal Structure Analysis of VP39 F180W mutant | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Oguro, A, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2001-08-20 | | Release date: | 2002-07-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The "cap-binding slot" of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap.
Biochemistry, 41, 2002
|
|
6LQV
 
 | |
6QZP
 
 | | High-resolution cryo-EM structure of the human 80S ribosome | | Descriptor: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 18S rRNA (1740-MER), 28S rRNA (3773-MER), ... | | Authors: | Natchiar, S.K, Myasnikov, A.G, Kratzat, H, Hazemann, I, Klaholz, B.P. | | Deposit date: | 2019-03-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualization of chemical modifications in the human 80S ribosome structure.
Nature, 551, 2017
|
|
