2Q4A
 
 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At3g21360 | | Descriptor: | Clavaminate synthase-like protein At3g21360, FE (III) ION, SULFATE ION | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q4J
 
 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At3g03250, a putative UDP-glucose pyrophosphorylase | | Descriptor: | Probable UTP-glucose-1-phosphate uridylyltransferase 2 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
3N8E
 
 | | Substrate binding domain of the human Heat Shock 70kDa protein 9 (mortalin) | | Descriptor: | Stress-70 protein, mitochondrial | | Authors: | Wisniewska, M, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, van der Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate binding domain of the human Heat Shock 70kDa protein 9 (mortalin)
To be published
|
|
2Q4K
 
 | | Ensemble refinement of the protein crystal structure of gene product from Homo sapiens Hs.433573 | | Descriptor: | Uncharacterized protein C11orf68 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q4O
 
 | | Ensemble refinement of the crystal structure of a lysine decarboxylase-like protein from Arabidopsis thaliana gene At2g37210 | | Descriptor: | MAGNESIUM ION, SULFATE ION, Uncharacterized protein At2g37210/T2N18.3 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q4Z
 
 | | Ensemble refinement of the protein crystal structure of an aspartoacylase from Rattus norvegicus | | Descriptor: | Aspartoacylase, SULFATE ION, ZINC ION | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q40
 
 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At2g17340 | | Descriptor: | MAGNESIUM ION, Protein At2g17340 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q4V
 
 | | Ensemble refinement of the protein crystal structure of thialysine n-acetyltransferase (SSAT2) from Homo sapiens | | Descriptor: | ACETYL COENZYME *A, Diamine acetyltransferase 2 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q51
 
 | | Ensemble refinement of the protein crystal structure of an aspartoacylase from Homo sapiens | | Descriptor: | Aspartoacylase, PHOSPHATE ION, ZINC ION | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
1A56
 
 | | PRIMARY SEQUENCE AND SOLUTION CONFORMATION OF FERRICYTOCHROME C-552 FROM NITROSOMONAS EUROPAEA, NMR, MEAN STRUCTURE REFINED WITH EXPLICIT HYDROGEN BOND CONSTRAINTS | | Descriptor: | FERRICYTOCHROME C-552, HEME C | | Authors: | Timkovich, R, Bergmann, D, Arciero, D.M, Hooper, A.B. | | Deposit date: | 1998-02-20 | | Release date: | 1998-10-21 | | Last modified: | 2020-12-16 | | Method: | SOLUTION NMR | | Cite: | Primary sequence and solution conformation of ferrocytochrome c-552 from Nitrosomonas europaea.
Biophys.J., 75, 1998
|
|
2Q3W
 
 | | Ensemble refinement of the protein crystal structure of the cys84ala cys85ala double mutant of the [2Fe-2S] ferredoxin subunit of toluene-4-monooxygenase from Pseudomonas mendocina KR1 | | Descriptor: | 1,2-ETHANEDIOL, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, ... | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q3T
 
 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At3g22680 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Protein At3g22680, ... | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2O4W
 
 | | T4 lysozyme circular permutant | | Descriptor: | CHLORIDE ION, Lysozyme | | Authors: | Llinas, M, Crowder, S.M, Echols, N, Alber, T, Marqusee, S. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
1D5V
 
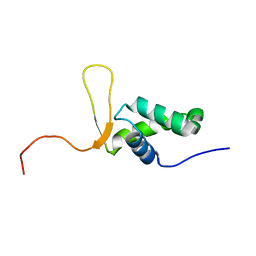 | | SOLUTION STRUCTURE OF THE FORKHEAD DOMAIN OF THE ADIPOCYTE-TRANSCRIPTION FACTOR FREAC-11 (S12) | | Descriptor: | S12 TRANSCRIPTION FACTOR (FKH-14) | | Authors: | van Dongen, M.J.P, Cederberg, A, Carlsson, P, Enerback, S, Wikstrom, M. | | Deposit date: | 1999-10-12 | | Release date: | 2000-10-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the DNA-binding domain of the adipocyte-transcription factor FREAC-11.
J.Mol.Biol., 296, 2000
|
|
5KSH
 
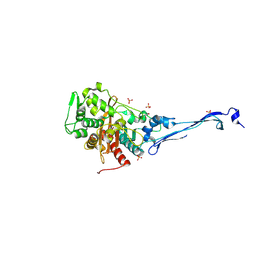 | |
5BRV
 
 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | Carbonic anhydrase 2, ZINC ION, pentamethylcyclopentadienyl iridium [N-benzensulfonamide-(2-pyridylmethyl-4-benzensulfonamide)amin] chloride | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
6E9N
 
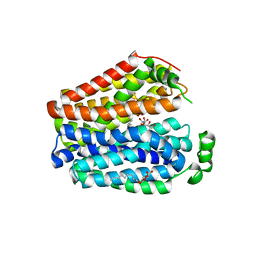 | |
6E9O
 
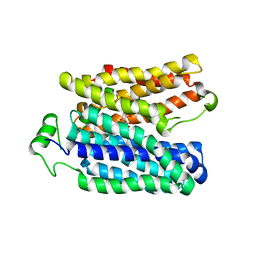 | |
6J3Z
 
 | | Structure of C2S1M1-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
6J3Y
 
 | | Structure of C2S2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
6J40
 
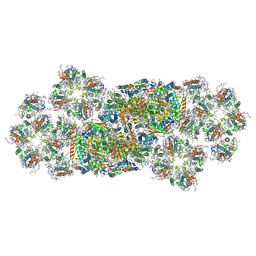 | | Structure of C2S2M2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
2JOL
 
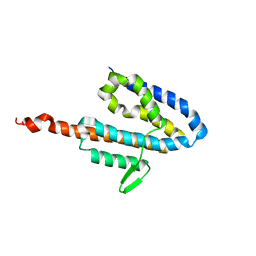 | | Average NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | Descriptor: | Putative G-nucleotide exchange factor | | Authors: | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-03-14 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
7QZN
 
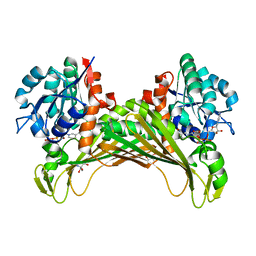 | | Amine Dehydrogenase from Cystobacter fuscus (CfusAmDH) W145A mutant with NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amine Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Bennett, M, Ducrot, L, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Cover Feature: Expanding the Substrate Scope of Native Amine Dehydrogenases through In Silico Structural Exploration and Targeted Protein Engineering (ChemCatChem 22/2022)
Chemcatchem, 14, 2022
|
|
7QZL
 
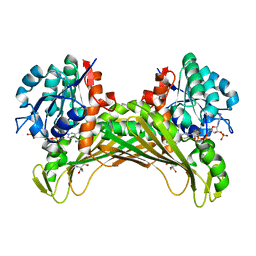 | | Amine Dehydrogenase from Cystobacter fuscus (CfusAmDH) W145A mutant with NADP+ and pentylamine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AMYLAMINE, Amine Dehydrogenase, ... | | Authors: | Bennett, M, Ducrot, L, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cover Feature: Expanding the Substrate Scope of Native Amine Dehydrogenases through In Silico Structural Exploration and Targeted Protein Engineering (ChemCatChem 22/2022)
Chemcatchem, 14, 2022
|
|
5E4D
 
 | |
