7AZ2
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1014 | | Descriptor: | 1-[4-methyl-2-(trifluoromethyl)phenyl]-2-phenyl-imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-14 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.081 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7AOG
 
 | | 14-3-3 sigma in complex with Pin1 binding site pS72 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7AZ1
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1013 | | Descriptor: | 1-[4-methyl-3-(trifluoromethyl)phenyl]-2-phenyl-imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-14 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7ZMW
 
 | |
7ZMU
 
 | |
4RN1
 
 | | Crystal structure of S39D HDAC8 in complex with a largazole analogue. | | Descriptor: | (5R,8S,11S)-5-methyl-8-(propan-2-yl)-11-[(1E)-4-sulfanylbut-1-en-1-yl]-3-thia-7,10,14,20,21-pentaazatricyclo[14.3.1.1~2,5~]henicosa-1(20),2(21),16,18-tetraene-6,9,13-trione, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Variable Active Site Loop Conformations Accommodate the Binding of Macrocyclic Largazole Analogues to HDAC8.
Biochemistry, 54, 2015
|
|
1JWH
 
 | | Crystal Structure of Human Protein Kinase CK2 Holoenzyme | | Descriptor: | Casein kinase II beta chain, Casein kinase II, alpha chain, ... | | Authors: | Niefind, K, Guerra, B, Ermakowa, I, Issinger, O.G. | | Deposit date: | 2001-09-04 | | Release date: | 2002-03-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of human protein kinase CK2: insights into basic properties of the CK2 holoenzyme.
EMBO J., 20, 2001
|
|
7ZTD
 
 | | Non-muscle F-actin decorated with non-muscle tropomyosin 3.2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Non-muscle tropomyosin 3.2, actin, ... | | Authors: | Selvaraj, M, Kokate, S, Kogan, K, Kotila, T, Kremneva, E, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2022-05-09 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis underlying specific biochemical activities of non-muscle tropomyosin isoforms.
Cell Rep, 42, 2023
|
|
7ZTC
 
 | | Non-muscle F-actin decorated with non-muscle tropomyosin 1.6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Non-muscle tropomyosin 1.6, actin, ... | | Authors: | Selvaraj, M, Kokate, S, Kogan, K, Kotila, T, Kremneva, E, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2022-05-09 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis underlying specific biochemical activities of non-muscle tropomyosin isoforms.
Cell Rep, 42, 2023
|
|
4RN0
 
 | | Crystal structure of S39D HDAC8 in complex with a largazole analogue. | | Descriptor: | (5R,8S,11S)-5-methyl-8-(propan-2-yl)-11-[(1E)-4-sulfanylbut-1-en-1-yl]-3,17-dithia-7,10,14,19,20-pentaazatricyclo[14.2.1.1~2,5~]icosa-1(18),2(20),16(19)-triene-6,9,13-trione, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Variable Active Site Loop Conformations Accommodate the Binding of Macrocyclic Largazole Analogues to HDAC8.
Biochemistry, 54, 2015
|
|
2H3B
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor 1 | | Descriptor: | Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4J4L
 
 | |
9ET8
 
 | | CDK2-cyclin A in complex with FragLite 5 | | Descriptor: | 4-iodanylpyridin-2-amine, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Hope, I, Martin, M.P, Waring, M.J, Noble, M.E.M, Endicott, J.A, Tatum, N.J. | | Deposit date: | 2024-03-26 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystallographic fragment screening of CDK2-cyclin A: FragLites map sites of protein-protein interaction
To Be Published
|
|
4I77
 
 | | Lebrikizumab Fab bound to IL-13 | | Descriptor: | Interleukin-13, Lebrikizumab heavy chain, Lebrikizumab light chain | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Signaling Blockade by Anti-IL-13 Antibody Lebrikizumab.
J.Mol.Biol., 425, 2013
|
|
3A03
 
 | | Crystal structure of Hox11L1 homeodomain | | Descriptor: | SODIUM ION, SULFATE ION, T-cell leukemia homeobox protein 2 | | Authors: | Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2009-02-28 | | Release date: | 2010-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
3A02
 
 | | Crystal structure of Aristaless homeodomain | | Descriptor: | CADMIUM ION, CHLORIDE ION, Homeobox protein aristaless | | Authors: | Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2009-02-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
3A01
 
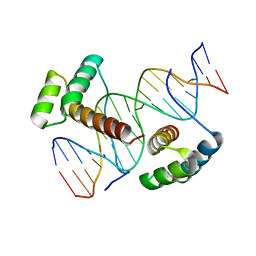 | | Crystal structure of Aristaless and Clawless homeodomains bound to DNA | | Descriptor: | 5'-D(*CP*CP*GP*CP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*GP*CP*GP*G)-3', Homeobox protein aristaless, ... | | Authors: | Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2009-02-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
1YSO
 
 | | YEAST CU, ZN SUPEROXIDE DISMUTASE WITH THE REDUCED BRIDGE BROKEN | | Descriptor: | COPPER (I) ION, YEAST CU, ZN SUPEROXIDE DISMUTASE, ... | | Authors: | Parge, H.E, Crane, B.R, Tsang, J, Tainer, J.A. | | Deposit date: | 1995-12-21 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Unusual trigonal-planar copper configuration revealed in the atomic structure of yeast copper-zinc superoxide dismutase.
Biochemistry, 35, 1996
|
|
1WCC
 
 | | Screening for fragment binding by X-ray crystallography | | Descriptor: | 2-AMINO-6-CHLOROPYRAZINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Cleasby, A, O'Reilly, M, Hartshorn, M.J, Murray, C.W, Tickle, I.J, Jhoti, H, Frederickson, M. | | Deposit date: | 2004-11-12 | | Release date: | 2005-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
5VPC
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-II crystal | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M.C, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPF
 
 | | Transcription factor FosB/JunD bZIP domain in complex with cognate DNA, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*TP*CP*GP*GP*TP*GP*AP*CP*TP*CP*AP*CP*CP*GP*AP*CP*G)-3'), ... | | Authors: | Yin, Z, Rudenko, G, Machius, M. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
1GNC
 
 | |
2R6M
 
 | | Crystal structure of rat CK2-beta subunit | | Descriptor: | Casein kinase II subunit beta, ZINC ION | | Authors: | Shen, Y. | | Deposit date: | 2007-09-06 | | Release date: | 2008-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of catalytic and regulatory subunits of rat protein kinase CK2
CHIN.SCI.BULL., 54, 2009
|
|
1K6X
 
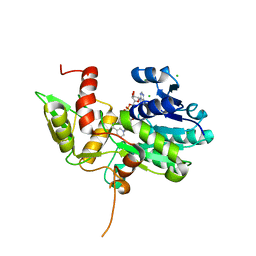 | | Crystal structure of Nmra, a negative transcriptional regulator in complex with NAD at 1.5 A resolution (Trigonal form) | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2002
|
|
2H3D
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor in Complex with Nicotinamide Mononuleotide | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide phosphoribosyltransferase | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
