9BTM
 
 | | NRas 1-169 Q61R in Complex with Shoc2 80-582 | | Descriptor: | GTPase NRas, GUANOSINE-5'-TRIPHOSPHATE, Leucine-rich repeat protein SHOC-2, ... | | Authors: | Hauseman, Z.J, King, D, Viscomi, J, Fodor, M. | | Deposit date: | 2024-05-15 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Targeting the SHOC2-RAS interaction in RAS-mutant cancers.
Nature, 2025
|
|
3VCH
 
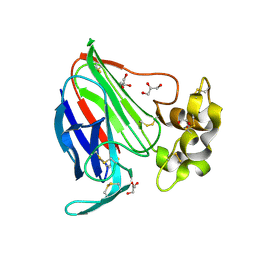 | | Thaumatin by Classical Hanging Drop Vapour Diffusion after 9.05 MGy X-Ray dose at ESRF ID29 beamline (Best Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Pechkova, E, Scudieri, D, Nicolini, C. | | Deposit date: | 2012-01-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
Crit Rev Eukaryot Gene Expr, 22, 2012
|
|
6EV5
 
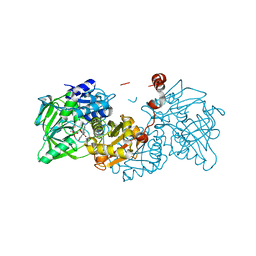 | | Crystal structure of E282Q A. niger Fdc1 with prFMN in the hydroxylated form | | Descriptor: | Ferulic acid decarboxylase 1, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Bailey, S.S, Leys, D, Payne, K.A.P. | | Deposit date: | 2017-11-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The role of conserved residues in Fdc decarboxylase in prenylated flavin mononucleotide oxidative maturation, cofactor isomerization, and catalysis.
J. Biol. Chem., 293, 2018
|
|
7AYN
 
 | | Crystal structure of the lectin domain of the FimH variant Arg98Ala, in complex with Methyl 3-chloro-4-D-mannopyranosyloxy-3-biphenylcarboxylate | | Descriptor: | 1,2-ETHANEDIOL, Type 1 fimbrin D-mannose specific adhesin, methyl 3-[3-chloranyl-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-phenyl]benzoate | | Authors: | Jakob, R.P, Tomasic, T, Rabbani, S, Reisner, A, Jakopin, Z, Maier, T, Ernst, B, Anderluh, M. | | Deposit date: | 2020-11-12 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Does targeting Arg98 of FimH lead to high affinity antagonists?
Eur.J.Med.Chem., 211, 2020
|
|
2ID9
 
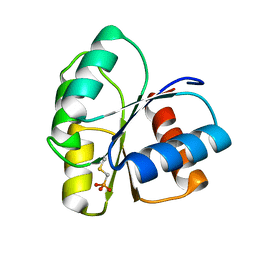 | | 1.85 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
5KSW
 
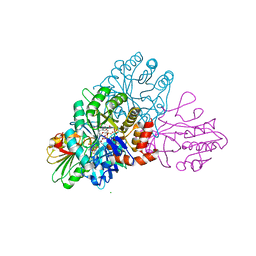 | | DHODB-I74D mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dihydroorotate dehydrogenase, ... | | Authors: | Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2016-07-10 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Isoleucine 74 Plays a Key Role in Controlling Electron Transfer Between Lactococcus lactis Dihydroorotate Dehydrogenase 1B Subunits
To Be Published
|
|
5H9Q
 
 | | Crystal Structure of Human Galectin-7 in Complex with TD139 | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, Galectin-7 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
6BDO
 
 | |
1KI0
 
 | | The X-ray Structure of Human Angiostatin | | Descriptor: | ANGIOSTATIN, BICINE | | Authors: | Abad, M.C, Arni, R.K, Grella, D.K, Castellino, F.J, Tulinsky, A, Geiger, J.H. | | Deposit date: | 2001-12-02 | | Release date: | 2002-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray crystallographic structure of the angiogenesis inhibitor angiostatin.
J.Mol.Biol., 318, 2002
|
|
1KHM
 
 | | C-TERMINAL KH DOMAIN OF HNRNP K (KH3) | | Descriptor: | PROTEIN (HNRNP K) | | Authors: | Baber, J, Libutti, D, Levens, D, Tjandra, N. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K, a c-myc transcription factor.
J.Mol.Biol., 289, 1999
|
|
7YAA
 
 | | Crystal structure analysis of cp3 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, GLYCEROL, N-(2-acetamidoethyl)-4-(4-methanoyl-1,3-thiazol-2-yl)benzamide, ... | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7Y8D
 
 | | Crystal structure of cp1 bound BCLxl | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Bcl-2-like protein 1, cp1 peptide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
5E0Q
 
 | |
5YCL
 
 | |
7YA5
 
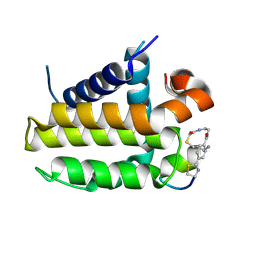 | | Crystal structure analysis of cp1 bound BCL2/G101V | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Apoptosis regulator Bcl-2, cp1 peptide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
4ZGC
 
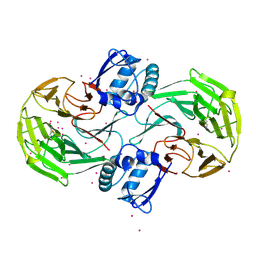 | | Crystal Structure Analysis of Kelch protein (with disulfide bond) from Plasmodium falciparum | | Descriptor: | Kelch protein, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Ravichandran, M, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, El Bakkouri, M, Senisterra, G, Osman, K.T, Lovato, D.V, Hui, R, Hutchinson, A, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-04-22 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of kelch protein with disulfide bond from Plasmodium falciparum.
to be published
|
|
1KQH
 
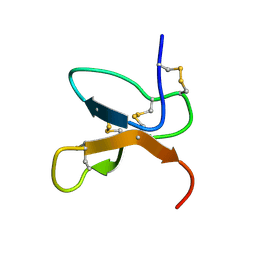 | | NMR Solution Structure of the cis Pro30 Isomer of ACTX-Hi:OB4219 | | Descriptor: | ACTX-Hi:OB4219 | | Authors: | Rosengren, K.J, Wilson, D, Daly, N.L, Alewood, P.F, Craik, D.J. | | Deposit date: | 2002-01-05 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the cis- and trans-Pro30 isomers of a novel 38-residue toxin
from the venom of Hadronyche Infensa sp. that contains a cystine-knot motif within
its four disulfide bonds
Biochemistry, 41, 2002
|
|
3W1M
 
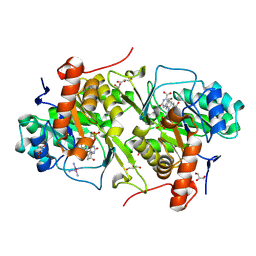 | | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with 5-bromoorotate | | Descriptor: | 5-bromo-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with 5-bromoorotate
To be Published
|
|
1KQI
 
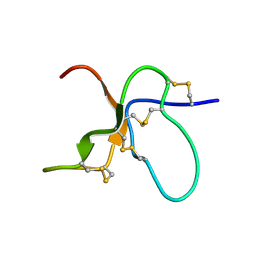 | | NMR Solution Structure of the trans Pro30 Isomer of ACTX-Hi:OB4219 | | Descriptor: | ACTX-Hi:OB4219 | | Authors: | Rosengren, K.J, Wilson, D, Daly, N.L, Alewood, P.F, Craik, D.J. | | Deposit date: | 2002-01-06 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the cis- and trans-Pro30 isomers of a novel 38-residue toxin
from the venom of Hadronyche Infensa sp. that contains a cystine-knot motif within
its four disulfide bonds
Biochemistry, 41, 2002
|
|
6ITS
 
 | |
4P7Y
 
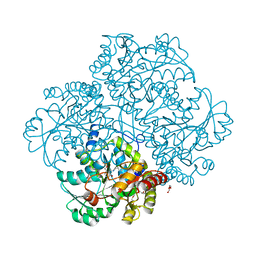 | | L-methionine gamma-lyase from Citrobacter freundii with Y58F substitution | | Descriptor: | DI(HYDROXYETHYL)ETHER, METHIONINE GAMMA-LYASE, PENTAETHYLENE GLYCOL | | Authors: | Revtovich, S.V, Nikulin, A.D, Anufrieva, N.V, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2014-03-28 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of L-methionine gamma-lyase from Citrobacter freundii with Y58F substitution
To Be Published
|
|
2IL9
 
 | |
9F59
 
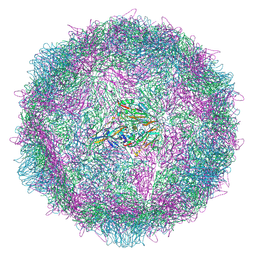 | | Poliovirus type 2 (strain MEF-1) stabilised virus-like particle (PV2 SC6b) from a mammalian expression system. | | Descriptor: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3, ... | | Authors: | Bahar, M.W, Porta, C, Fry, E.E, Stuart, D.I. | | Deposit date: | 2024-04-28 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Recombinant expression systems for production of stabilised virus-like particles as next-generation polio vaccines.
Nat Commun, 16, 2025
|
|
5LAV
 
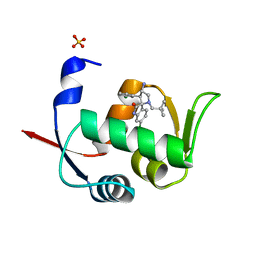 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) in complex with compound 6b | | Descriptor: | (3~{S},3'~{S},4'~{S})-4'-azanyl-6-chloranyl-3'-(3-chlorophenyl)-1'-(2,2-dimethylpropyl)spiro[1~{H}-indole-3,2'-pyrrolidine]-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
1KKO
 
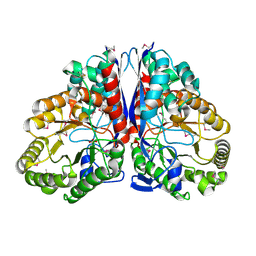 | | CRYSTAL STRUCTURE OF CITROBACTER AMALONATICUS METHYLASPARTATE AMMONIA LYASE | | Descriptor: | 3-METHYLASPARTATE AMMONIA-LYASE, SULFATE ION | | Authors: | Levy, C.W, Buckley, P.A, Sedelnikova, S, Kato, Y, Asano, Y, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Insights into enzyme evolution revealed by the structure of methylaspartate ammonia lyase.
Structure, 10, 2002
|
|
