4KWC
 
 | |
4MHB
 
 | | Structure of a putative reductase from Yersinia pestis | | Descriptor: | Putative aldo/keto reductase, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Rembert, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a putative reductase from Yersinia pestis
To be Published
|
|
4LHP
 
 | | Crystal Structure of Native FG41Malonate Semialdehyde Decarboxylase | | Descriptor: | FG41 Malonate Semialdehyde Decarboxylase, PHOSPHATE ION, SULFATE ION | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
4MLG
 
 | | Structure of RS223-Beta-xylosidase | | Descriptor: | Beta-xylosidase, CALCIUM ION, SULFATE ION | | Authors: | Jordan, D, Braker, J, Wagschal, K, Lee, C, Dubrovska, I, Anderson, S, Wawrzak, Z. | | Deposit date: | 2013-09-06 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of RS223-Beta-xylosidase
To be Published
|
|
4OHN
 
 | | Crystal structure of an ABC uptake transporter substrate binding protein from Streptococcus pneumoniae with Bound Histidine | | Descriptor: | ABC transporter substrate-binding protein, ACETATE ION, HISTIDINE | | Authors: | Brunzelle, J.S, Wawrzak, W, Yim, Y, Kudritska, M, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of an ABC uptake transporter substrate binding protein from Streptococcus pneumoniae with Bound Histidine
To be Published
|
|
4OMV
 
 | | Crystal Structure of a Putative Macrophage Growth Locus, subunit A From Francisella tularensis SCHU S4 | | Descriptor: | Macrophage growth locus, subunit A | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-27 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of a Putative Macrophage Growth Locus, subunit A From Francisella tularensis SCHU S4
TO BE PUBLISHED
|
|
3M8A
 
 | | Crystal Structure of Swine Flu Virus NS1 N-Terminal RNA Binding Domain from H1N1 Influenza A/California/07/2009 | | Descriptor: | ACETATE ION, MALONATE ION, Nonstructural protein 1, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-17 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Swine Flu Virus NS1 N-Terminal RNA Binding Domain from H1N1 Influenza A/California/07/2009
To be Published
|
|
3N84
 
 | |
3N08
 
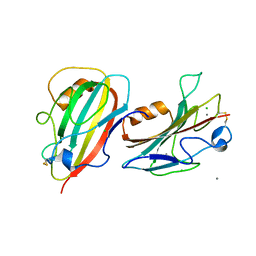 | | Crystal Structure of a Putative PhosphatidylEthanolamine-Binding Protein (PEBP) Homolog CT736 from Chlamydia trachomatis D/UW-3/CX | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative PhosphatidylEthanolamine-Binding Protein (PEBP) | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-13 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 Angstrom Crystal Structure of a Putative PhosphatidylEthanolamine-Binding Protein (PEBP) Homolog CT736 from Chlamydia trachomatis D/UW-3/CX
To be Published
|
|
3N0L
 
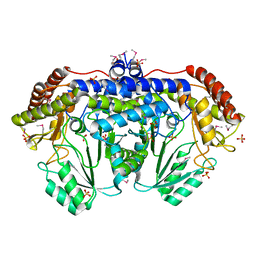 | | Crystal structure of serine hydroxymethyltransferase from Campylobacter jejuni | | Descriptor: | SULFATE ION, Serine hydroxymethyltransferase | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-14 | | Release date: | 2010-05-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of serine hydroxymethyltransferase from Campylobacter jejuni
TO BE PUBLISHED
|
|
3N7Y
 
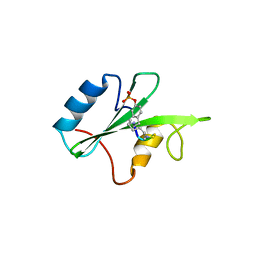 | |
3NEW
 
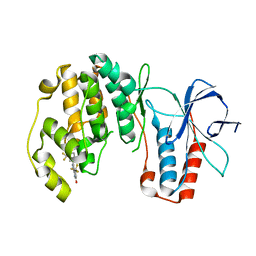 | | p38-alpha complexed with Compound 10 | | Descriptor: | 4-(trifluoromethyl)-3-[3-(trifluoromethyl)phenyl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, Mitogen-activated protein kinase 14 | | Authors: | Goedken, E.R, Comess, K.M, Sun, C, Argiriadi, M, Jia, Y, Quinn, C.M, Banach, D.L, Marcotte, D, Borhani, D. | | Deposit date: | 2010-06-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Characterization of Non-ATP Site Inhibitors of the Mitogen Activated Protein (MAP) Kinases.
Acs Chem.Biol., 6, 2011
|
|
6F0K
 
 | | Alternative complex III | | Descriptor: | ActD, ActF, ActH, ... | | Authors: | Sousa, J.S, Calisto, F, Mills, D.J, Pereira, M.M, Vonck, J, Kuehlbrandt, W. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural basis for energy transduction by respiratory alternative complex III.
Nat Commun, 9, 2018
|
|
3NX4
 
 | | Crystal structure of the yhdH oxidoreductase from Salmonella enterica in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Anderson, S.M, Wawrzak, Z, Peterson, S, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-12 | | Release date: | 2010-08-04 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the yhdH oxidoreductase from Salmonella enterica in complex with NADP
To be Published
|
|
3O04
 
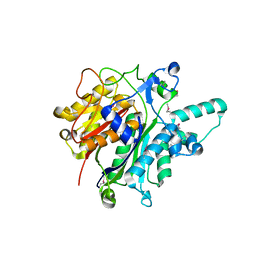 | | Crystal structure of the beta-keto-acyl carrier protein synthase II (lmo2201) from Listeria monocytogenes | | Descriptor: | beta-keto-acyl carrier protein synthase II | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-04 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the beta-keto-acyl carrier protein synthase II (lmo2201) from Listeria monocytogenes
To be Published
|
|
2VJ8
 
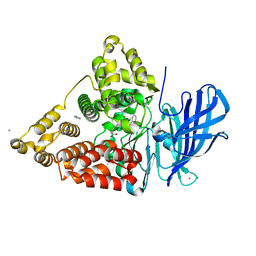 | | Complex of human leukotriene A4 hydrolase with a hydroxamic acid inhibitor | | Descriptor: | 6-[{(2S)-2-AMINO-3-[4-(BENZYLOXY)PHENYL]PROPYL}(HYDROXY)AMINO]-6-OXOHEXANOIC ACID), ACETATE ION, IMIDAZOLE, ... | | Authors: | Thunnissen, M.M.G.M, Andersson, B, Wong, C.-H, Samuelsson, B, Haeggstrom, J.Z. | | Deposit date: | 2007-12-07 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Leukotriene A4 Hydrolase in Complex with Captopril and Two Competitive Tight-Binding Inhibitors
Faseb J., 16, 2002
|
|
2WHR
 
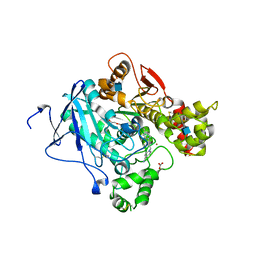 | | Crystal structure of acetylcholinesterase in complex with K027 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Ekstrom, F, Hornberg, A, Artursson, E, Hammarstrom, L.-G, Schneider, G, Pang, Y.-P. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Structure of Hi-6Sarin-Acetylcholinesterase Determined by X-Ray Crystallography and Molecular Dynamics Simulation: Reactivator Mechanism and Design.
Plos One, 4, 2009
|
|
2WNE
 
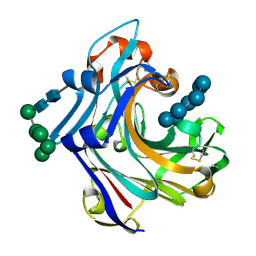 | | Mutant Laminarinase 16A cyclizes laminariheptaose | | Descriptor: | PUTATIVE LAMINARINASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Vasur, J, Kawai, R, Andersson, E, Widmalm, G, Jonsson, K.H.M, Hansson, H, Engstrom, A, Einarsson, E, Forsberg, Z, Igarashi, K, Sandgren, M, Samejima, M, Stahlberg, J. | | Deposit date: | 2009-07-09 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | Synthesis of Cyclic Beta-Glucan Using Laminarinase 16A Glycosynthase Mutant from the Basidiomycete Phanerochaete Chrysosporium.
J.Am.Chem.Soc., 132, 2010
|
|
3OD6
 
 | | Crystal structure of p38alpha Y323T active mutant | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Livnah, O, Tzarum, N. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.682 Å) | | Cite: | Active mutants of the TCR-mediated p38alpha alternative activation site show changes in the phosphorylation lip and DEF site formation.
J.Mol.Biol., 405, 2011
|
|
6F36
 
 | | Polytomella Fo model | | Descriptor: | Mitochondrial ATP synthase subunit 6, Mitochondrial ATP synthase subunit ASA6, Mitochondrial ATP synthase subunit c | | Authors: | Yildiz, O, Kuehlbrandt, W, Klusch, N, Murphy, B.J, Mills, D.J. | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of proton translocation and force generation in mitochondrial ATP synthase.
Elife, 6, 2017
|
|
2X41
 
 | |
6FHZ
 
 | |
6F0E
 
 | | Structure of yeast Sec14p with a picolinamide compound | | Descriptor: | SEC14 cytosolic factor, ~{N}-(1,3-benzodioxol-5-ylmethyl)-5-bromanyl-3-fluoranyl-pyridine-2-carboxamide | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Target Identification and Mechanism of Action of Picolinamide and Benzamide Chemotypes with Antifungal Properties.
Cell Chem Biol, 25, 2018
|
|
3OCG
 
 | | P38 Alpha kinase complexed with a 5-amino-pyrazole based inhibitor | | Descriptor: | 5-amino-N-[5-(isoxazol-3-ylcarbamoyl)-2-methylphenyl]-1-phenyl-1H-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-08-10 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 5-Amino-pyrazoles as potent and selective p38α inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2X42
 
 | |
