3TVD
 
 | | Crystal Structure of Mouse RhoA-GTP complex | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Swaminathan, K, Pal, K, Jobichen, C. | | Deposit date: | 2011-09-20 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Crystal structure of mouse RhoA:GTPgammaS complex in a centered lattice.
J.Struct.Funct.Genom., 13, 2012
|
|
4HKC
 
 | |
3LNQ
 
 | | Structure of Aristaless homeodomain in complex with DNA | | Descriptor: | 5'-D(*CP*CP*CP*TP*AP*AP*TP*TP*AP*AP*AP*CP*CP*C)-3', 5'-D(*GP*GP*GP*TP*TP*TP*AP*AP*TP*TP*AP*GP*GP*G)-3', ACETATE ION, ... | | Authors: | Takamura, Y, Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
1YWT
 
 | | Crystal structure of the human sigma isoform of 14-3-3 in complex with a mode-1 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, synthetic optimal phosphopeptide (mode-1) | | Authors: | Wilker, E.W, Grant, R.A, Artim, S.C, Yaffe, M.B. | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for 14-3-3sigma functional specificity.
J.Biol.Chem., 280, 2005
|
|
3LQ5
 
 | | Structure of CDK9/CyclinT in complex with S-CR8 | | Descriptor: | (2S)-2-({9-(1-methylethyl)-6-[(4-pyridin-2-ylbenzyl)amino]-9H-purin-2-yl}amino)butan-1-ol, Cell division protein kinase 9, Cyclin-T1 | | Authors: | Hole, A.J, Endicott, J.A, Baumli, S. | | Deposit date: | 2010-02-08 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | CDK Inhibitors Roscovitine and CR8 Trigger Mcl-1 Down-Regulation and Apoptotic Cell Death in Neuroblastoma Cells
Genes Cancer, 1, 2010
|
|
1MD4
 
 | | A folding mutant of human class pi glutathione transferase, created by mutating glycine 146 of the wild-type protein to valine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, pi glutathione transferase | | Authors: | Kong, G.K.-W, Dragani, B, Aceto, A, Cocco, R, Mannervik, B, Stenberg, G, McKinstry, W.J, Polekhina, G, Parker, M.W. | | Deposit date: | 2002-08-06 | | Release date: | 2002-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contribution of Glycine 146 to a Conserved Folding Module Affecting Stability and Refolding of Human Glutathione Transferase P1-1
J.Biol.Chem., 278, 2003
|
|
7ZKE
 
 | | Mot1:TBP:DNA - pre-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (36-MER), ... | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6HG9
 
 | |
7Z8S
 
 | | Mot1:TBP:DNA - post hydrolysis state | | Descriptor: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-03-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z7N
 
 | | Mot1E1434Q:TBP:DNA - substrate recognition state | | Descriptor: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZB5
 
 | | Mot1(1-1836):TBP:DNA - post-hydrolysis complex dimer | | Descriptor: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-03-23 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1MD3
 
 | | A folding mutant of human class pi glutathione transferase, created by mutating glycine 146 of the wild-type protein to alanine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, pi glutathione transferase | | Authors: | Kong, G.K.-W, Dragani, B, Aceto, A, Cocco, R, Mannervik, B, Stenberg, G, McKinstry, W.J, Polekhina, G, Parker, M.W. | | Deposit date: | 2002-08-06 | | Release date: | 2002-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Contribution of Glycine 146 to a Conserved Folding Module Affecting Stability and Refolding of Human Glutathione Transferase P1-1
J.Biol.Chem., 278, 2003
|
|
5CFU
 
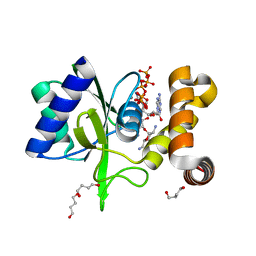 | | Crystal Structure of ANT(2")-Ia in complex with adenylyl-2"-tobramycin | | Descriptor: | 1,4-BUTANEDIOL, Aminoglycoside Nucleotidyltransferase (2")-Ia, MANGANESE (II) ION, ... | | Authors: | Rodionov, D, Bassenden, A.V, Berghuis, A.M. | | Deposit date: | 2015-07-08 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Revisiting the Catalytic Cycle and Kinetic Mechanism of AminoglycosideO-Nucleotidyltransferase(2′′): A Structural and Kinetic Study.
Acs Chem.Biol., 2020
|
|
2CCI
 
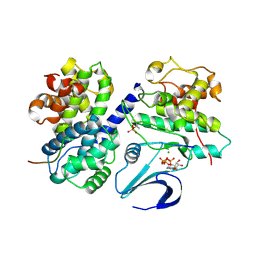 | | Crystal structure of phospho-CDK2 Cyclin A in complex with a peptide containing both the substrate and recruitment sites of CDC6 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6 homolog, Cyclin-A2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition.
J. Biol. Chem., 281, 2006
|
|
1JY0
 
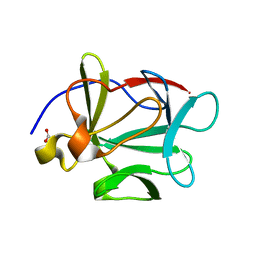 | |
2UUE
 
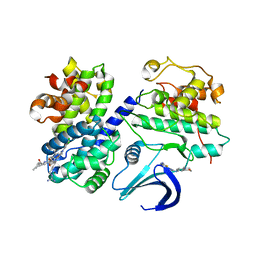 | | REPLACE: A strategy for Iterative Design of Cyclin Binding Groove Inhibitors | | Descriptor: | 1-(3,5-DICHLOROPHENYL)-5-METHYL-1H-1,2,4-TRIAZOLE-3-CARBOXYLIC ACID, 4-METHYL-5-{(2E)-2-[(4-MORPHOLIN-4-YLPHENYL)IMINO]-2,5-DIHYDROPYRIMIDIN-4-YL}-1,3-THIAZOL-2-AMINE, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Andrews, M.J, Kontopidis, G, McInnes, C, Plater, A, Innes, L, Cowan, A, Jewsbury, P, Fischer, P.M. | | Deposit date: | 2007-03-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Replace: A Strategy for Iterative Design of Cyclin- Binding Groove Inhibitors
Chembiochem, 7, 2006
|
|
2PF4
 
 | |
4EON
 
 | | Thr 160 phosphorylated CDK2 H84S, Q85M, Q131E - human cyclin A3 complex with the inhibitor RO3306 | | Descriptor: | (5E)-5-(quinolin-6-ylmethylidene)-2-[(thiophen-2-ylmethyl)amino]-1,3-thiazol-4(5H)-one, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4EOJ
 
 | | Thr 160 phosphorylated CDK2 H84S, Q85M, K89D - human cyclin A3 complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4EOK
 
 | | Thr 160 phosphorylated CDK2 H84S, Q85M, K89D - human cyclin A3 complex with the inhibitor NU6102 | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, MONOTHIOGLYCEROL, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4EOS
 
 | | Thr 160 phosphorylated CDK2 WT - human cyclin A3 complex with the inhibitor RO3306 | | Descriptor: | (5E)-5-(quinolin-6-ylmethylidene)-2-[(thiophen-2-ylmethyl)amino]-1,3-thiazol-4(5H)-one, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
2F2C
 
 | | X-ray structure of human CDK6-Vcyclinwith the inhibitor aminopurvalanol | | Descriptor: | (2S)-2-({6-[(3-AMINO-5-CHLOROPHENYL)AMINO]-9-ISOPROPYL-9H-PURIN-2-YL}AMINO)-3-METHYLBUTAN-1-OL, Cell division protein kinase 6, Cyclin homolog, ... | | Authors: | Schulze-Gahmen, U, Lu, H. | | Deposit date: | 2005-11-16 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Toward understanding the structural basis of cyclin-dependent kinase 6 specific inhibition.
J.Med.Chem., 49, 2006
|
|
8SG2
 
 | | BIVALENT INTERACTIONS OF PIN1 WITH THE C-TERMINAL TAIL OF PKC | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, Protein kinase C beta type | | Authors: | Dixit, K, Yang, Y, Chen, X.R, Igumenova, T.I. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | A novel bivalent interaction mode underlies a non-catalytic mechanism for Pin1-mediated protein kinase C regulation.
Elife, 13, 2024
|
|
2CCH
 
 | | The crystal structure of CDK2 cyclin A in complex with a substrate peptide derived from CDC modified with a gamma-linked ATP analogue | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION CONTROL PROTEIN 6 HOMOLOG, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Cheng, K.Y, Noble, M.E.M, Skamnaki, V, Brown, N.R, Lowe, E.D, Kontogiannis, L, Shen, K, Cole, P.A, Siligardi, G, Johnson, L.N. | | Deposit date: | 2006-01-16 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Role of the Phospho-Cdk2/Cyclin a Recruitment Site in Substrate Recognition
J.Biol.Chem., 281, 2006
|
|
3QZT
 
 | | Crystal Structure of BPTF bromo in complex with histone H4K16ac - Form II | | Descriptor: | GLYCEROL, Histone H4, Nucleosome-remodeling factor subunit BPTF | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
