6RGZ
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 6.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RH8
 
 | | Revisiting pH-gated conformational switch. Complex HK853 mutant H260A -RR468 mutant D53A pH 5.3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-19 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RGY
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 7.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RFV
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Response regulator, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6S1K
 
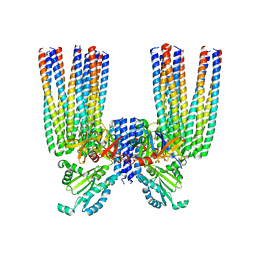 | | E. coli Core Signaling Unit, carrying QQQQ receptor mutation | | Descriptor: | CheW, Chemotaxis protein CheA, Methyl-accepting chemotaxis protein I | | Authors: | Cassidy, C.K. | | Deposit date: | 2019-06-18 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.38 Å) | | Cite: | Structure and dynamics of the E. coli chemotaxis core signaling complex by cryo-electron tomography and molecular simulations.
Commun Biol, 3, 2020
|
|
6RKW
 
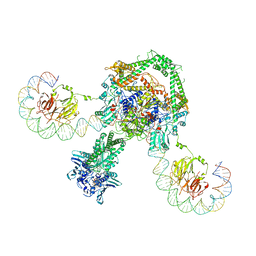 | | CryoEM structure of the complete E. coli DNA Gyrase complex bound to a 130 bp DNA duplex | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA (58-MER), DNA (62-MER), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
6RH2
 
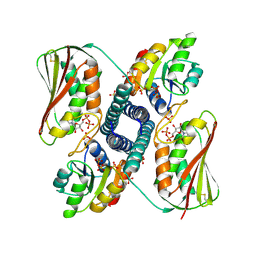 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 D53A pH 5.3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RKS
 
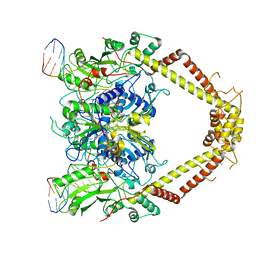 | | E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 without TOPRIM insertion | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA Strand 1, DNA Strand 2, ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
6RH7
 
 | | Revisiting pH-gated conformational switch. Complex HK853 mutant H260A -RR468 mutant D53A pH 7.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RKV
 
 | |
6RKU
 
 | | E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA Strand 1, DNA Strand 2, ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
6RH0
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 5.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Response regulator, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RH1
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 D53A pH 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
3A0R
 
 | | Crystal structure of histidine kinase ThkA (TM1359) in complex with response regulator protein TrrA (TM1360) | | Descriptor: | MERCURY (II) ION, Response regulator, Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3CRL
 
 | | Crystal structure of the PDHK2-L2 complex. | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial, MAGNESIUM ION, ... | | Authors: | Popov, K.M, Luo, M, Green, T.J, Grigorian, A, Klyuyeva, A, Tuganova, A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-04-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and functional insights into the molecular mechanisms responsible for the regulation of pyruvate dehydrogenase kinase 2.
J.Biol.Chem., 283, 2008
|
|
1ZXM
 
 | | Human Topo IIa ATPase/AMP-PNP | | Descriptor: | DNA topoisomerase II, alpha isozyme, MAGNESIUM ION, ... | | Authors: | Wei, H, Ruthenburg, A.J, Bechis, S.K, Verdine, G.L. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Nucleotide-dependent Domain Movement in the ATPase Domain of a Human Type IIA DNA Topoisomerase.
J.Biol.Chem., 280, 2005
|
|
1B3Q
 
 | | CRYSTAL STRUCTURE OF CHEA-289, A SIGNAL TRANSDUCING HISTIDINE KINASE | | Descriptor: | MERCURY (II) ION, PROTEIN (CHEMOTAXIS PROTEIN CHEA) | | Authors: | Bilwes, A.M, Alex, L.A, Crane, B.R, Simon, M.I. | | Deposit date: | 1998-12-14 | | Release date: | 1999-12-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of CheA, a signal-transducing histidine kinase.
Cell(Cambridge,Mass.), 96, 1999
|
|
2C2A
 
 | |
1ZXN
 
 | | Human DNA topoisomerase IIa ATPase/ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA topoisomerase II, alpha isozyme, ... | | Authors: | Wei, H, Ruthenburg, A.J, Bechis, S.K, Verdine, G.L. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Nucleotide-dependent Domain Movement in the ATPase Domain of a Human Type IIA DNA Topoisomerase.
J.Biol.Chem., 280, 2005
|
|
3ZKB
 
 | | CRYSTAL STRUCTURE OF THE ATPASE REGION OF Mycobacterium tuberculosis GyrB WITH AMPPNP | | Descriptor: | DNA GYRASE SUBUNIT B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Agrawal, A, Roue, M, Spitzfaden, C, Petrella, S, Aubry, A, Volker, C, Mossakowska, D, Hann, M, Bax, B, Mayer, C. | | Deposit date: | 2013-01-22 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mycobacterium Tuberculosis DNA Gyrase ATPase Domain Structures Suggest a Dissociative Mechanism that Explains How ATP Hydrolysis is Coupled to Domain Motion.
Biochem.J., 456, 2013
|
|
7CCH
 
 | | Acinetobacter baumannii histidine kinase AdeS | | Descriptor: | AdeS | | Authors: | Wen, Y, Felix, J. | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.848 Å) | | Cite: | Proteolysis and multimerization regulate signaling along the two-component regulatory system AdeRS.
Iscience, 24, 2021
|
|
7CMP
 
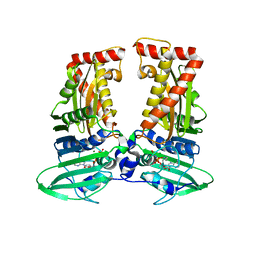 | | parE in complex with AMPPNP | | Descriptor: | DNA topoisomerase 4 subunit B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Jung, H.Y, Heo, Y.-S. | | Deposit date: | 2020-07-28 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Crystal structure of parE in complex with AMPPNP
To Be Published
|
|
4BIZ
 
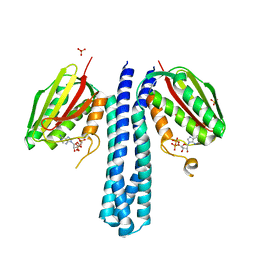 | |
4BIU
 
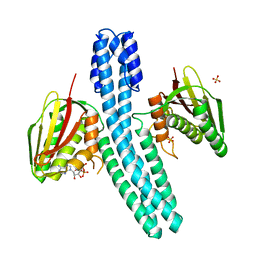 | | Crystal structure of CpxAHDC (orthorhombic form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SENSOR PROTEIN CPXA, SULFATE ION | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BIV
 
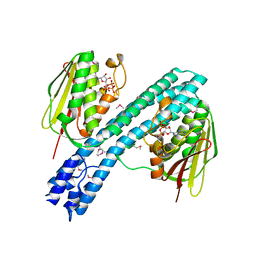 | | Crystal structure of CpxAHDC (trigonal form) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, SENSOR PROTEIN CPXA | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
