5L0S
 
 | | human POGLUT1 in complex with Factor VII EGF1 and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
6CJA
 
 | |
6XQH
 
 | | Crystal structure of SCLam E144S mutant, a non-specific endo-beta-1,3(4)-glucanase from family GH16, co-crystallized with cellotriose, presenting a 1,3-beta-D-cellobiosyl-glucose and a cellobiose at active site | | Descriptor: | CALCIUM ION, GH16 family protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Liberato, M.V, Squina, F. | | Deposit date: | 2020-07-09 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
7L5H
 
 | |
7PMT
 
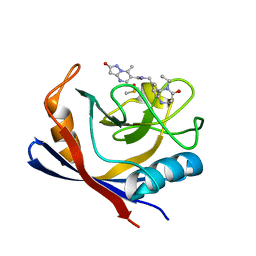 | | Human Cyclophilin D in complex with N-[(5-ethyl-4-oxo-1,2,3,4,5,6- hexahydro-1,5-benzodiazocin-8-yl)methyl]-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide | | Descriptor: | DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ... | | Authors: | Silva, D.O, Graedler, U, Bandeiras, T.M. | | Deposit date: | 2021-09-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Human Cyclophilin D in complex with N-[(4-aminophenyl)methyl]-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide
To Be Published
|
|
7ZMG
 
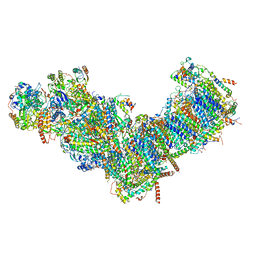 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
6RMX
 
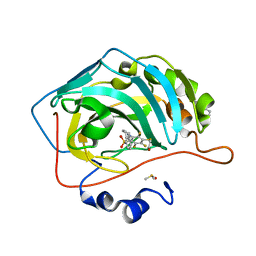 | | Human Carbonic anhydrase II mutant T199V bound by 2-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide | | Descriptor: | 2-[(1S)-2,3-dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)sulfanyl]benzenesulfonamide, BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Paketuryte, V, Manakova, E, Grazulis, S. | | Deposit date: | 2019-05-07 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human Carbonic anhydrase II mutant T199V bound by 2-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide
To Be Published
|
|
5W0I
 
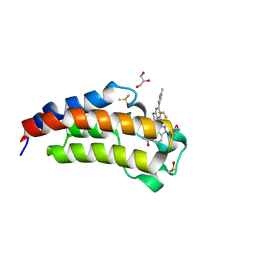 | | CREBBP Bromodomain in complex with Cpd8 (1-(3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydrofuran-3-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-[(3S)-oxolan-3-yl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-30 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
8QKZ
 
 | | Human Carbonic Anhydrase II in complex with 3,4-dihydro-1H-benzo[c][1,2]oxaborinin-1-ol at pH 9.0 | | Descriptor: | 1,1-bis(oxidanyl)-3,4-dihydro-2,1$l^{4}-benzoxaborinine, 1,2-ETHANEDIOL, Carbonic anhydrase 2, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-09-18 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Benzoxaborinine, New Chemotype for Carbonic Anhydrase Inhibition: Ex Novo Synthesis, Crystallography, In Silico Studies, and Anti-Melanoma Cell Line Activity.
J.Med.Chem., 67, 2024
|
|
8QHO
 
 | | Human Carbonic Anhydrase II in complex with 3,4-dihydro-1H-benzo[c][1,2]oxaborinin-1-ol | | Descriptor: | 1,1-bis(oxidanyl)-3,4-dihydro-2,1$l^{4}-benzoxaborinine, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-09-08 | | Release date: | 2024-09-18 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Benzoxaborinine, New Chemotype for Carbonic Anhydrase Inhibition: Ex Novo Synthesis, Crystallography, In Silico Studies, and Anti-Melanoma Cell Line Activity.
J.Med.Chem., 67, 2024
|
|
8CHS
 
 | |
8QQ9
 
 | | human carbonic anhydrase I in complex with 1-benzyl-3-(1-hydroxy-3,4-dihydro-1H-benzo[c][1,2]oxaborinin-7-yl)thiourea | | Descriptor: | 1-[1,1-bis(oxidanyl)-3,4-dihydro-2,1$l^{4}-benzoxaborinin-7-yl]-3-(phenylmethyl)thiourea, Carbonic anhydrase 1, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Benzoxaborinine, New Chemotype for Carbonic Anhydrase Inhibition: Ex Novo Synthesis, Crystallography, In Silico Studies, and Anti-Melanoma Cell Line Activity.
J.Med.Chem., 67, 2024
|
|
4Y0H
 
 | | Gamma-aminobutyric acid aminotransferase inactivated by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115) | | Descriptor: | 4-aminobutyrate aminotransferase, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rui, W, Ruslan, S, Hyunbeom, L, Emma, H.D, Jose, I.J, Neil, K, Richard, B.S, Dali, L. | | Deposit date: | 2015-02-06 | | Release date: | 2015-03-11 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mechanism of inactivation of gamma-aminobutyric acid aminotransferase by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115)
J. Am. Chem. Soc., 2015
|
|
3DMJ
 
 | | CRYSTAL STRUCTURE OF HIV-1 V106A and Y181C MUTANT REVERSE TRANSCRIPTASE IN COMPLEX WITH GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-07-01 | | Release date: | 2008-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
7LW3
 
 | | Structure of SARS-CoV-2 nsp16/nsp10 complex in presence of Cap-1 analog (m7GpppAmU) and SAH | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Gupta, Y.K, Viswanathan, T, Misra, A, Qi, S. | | Deposit date: | 2021-02-27 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A metal ion orients SARS-CoV-2 mRNA to ensure accurate 2'-O methylation of its first nucleotide.
Nat Commun, 12, 2021
|
|
6PQ8
 
 | | Crystal structure of TLA-1 S70G extended spectrum Beta-lactamase in complex with clavulanic acid | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ACETATE ION, ASPARTIC ACID, ... | | Authors: | Rudino-Pinera, E, Cifuentes-Castro, V.H, Rodriguez-Almazan, C. | | Deposit date: | 2019-07-08 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of ESBL TLA-1 in complex with clavulanic acid reveals a second acylation site.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
9C2K
 
 | |
8RLK
 
 | | Structure of the apo form of PIB-1 in an Orthorombic space group | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Class C beta-lactamase-related serine hydrolase, MAGNESIUM ION, ... | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new type of Class C beta-lactamases defined by PIB-1. A metal-dependent carbapenem-hydrolyzing beta-lactamase, from Pseudomonas aeruginosa: Structural and functional analysis.
Int.J.Biol.Macromol., 277, 2024
|
|
3DLE
 
 | |
3DM2
 
 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
8HE8
 
 | | Human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-(3-oxidanylidene-4-pentan-3-yl-piperazin-1-yl)carbonyl-phenyl]methyl]quinazoline-2,4-dione, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Xu, B.L, Zhou, J. | | Deposit date: | 2022-11-07 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of Quinazoline-2,4(1 H ,3 H )-dione Derivatives Containing a Piperizinone Moiety as Potent PARP-1/2 Inhibitors─Design, Synthesis, In Vivo Antitumor Activity, and X-ray Crystal Structure Analysis.
J.Med.Chem., 66, 2023
|
|
3DI6
 
 | | HIV-1 RT with pyridazinone non-nucleoside inhibitor | | Descriptor: | 6-(4-chloro-2-fluoro-3-phenoxybenzyl)pyridazin-3(2H)-one, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Harris, S.F, Villasenor, A, Dunten, P. | | Deposit date: | 2008-06-19 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and optimization of pyridazinone non-nucleoside inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6SDL
 
 | | Human Carbonic Anhydrase II in complex with (R)-N-(3-(1H-indol-1-yl)-2-methylpropyl)-4-sulfamoylbenzamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, (R)-N-(3-INDOL-1-YL-2-METHYL-PROPYL)-4-SULFAMOYL-BENZAMIDE, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Gerber, H.D, Heine, A, Klebe, G. | | Deposit date: | 2019-07-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Human Carbonic Anhydrase II in complex with (R)-N-(3-(1H-indol-1-yl)-2-methylpropyl)-4-sulfamoylbenzamide
To Be Published
|
|
8REK
 
 | |
6D52
 
 | |
