4PIR
 
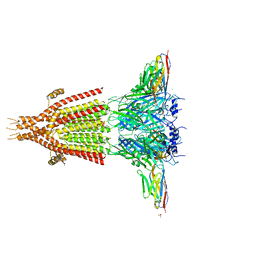 | | X-ray structure of the mouse serotonin 5-HT3 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Hassaine, G, Deluz, C, Grasso, L, Wyss, R, Tol, M.B, Hovius, R, Graff, A, Stahlberg, H, Tomizaki, T, Desmyter, A, Moreau, C, Li, X.-D, Poitevin, F, Vogel, H, Nury, H. | | Deposit date: | 2014-05-09 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | X-ray structure of the mouse serotonin 5-HT3 receptor.
Nature, 512, 2014
|
|
4PJT
 
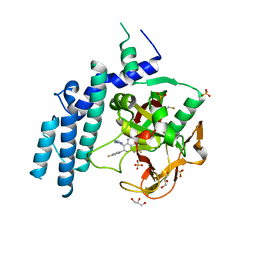 | | Structure of PARP1 catalytic domain bound to inhibitor BMN 673 | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, GLYCEROL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Aoyagi-Scharber, M, Gardberg, A.S, Arakaki, T.L. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the inhibition of poly(ADP-ribose) polymerases 1 and 2 by BMN 673, a potent inhibitor derived from dihydropyridophthalazinone.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
7NR1
 
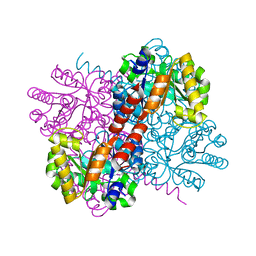 | | Crystal structure of holo-S116A mutant of Hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1 | | Descriptor: | HpcH/HpaI aldolase, MAGNESIUM ION | | Authors: | Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-03-02 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6YKR
 
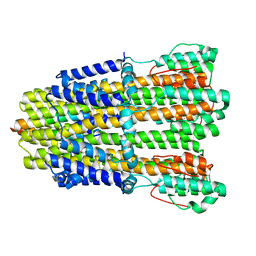 | |
4PFH
 
 | | Crystal structure of engineered D-tagatose 3-epimerase PcDTE-IDF8 | | Descriptor: | D-fructose, D-psicose, D-tagatose 3-epimerase, ... | | Authors: | Hee, C.S, Bosshart, A, Schirmer, T. | | Deposit date: | 2014-04-29 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed Divergent Evolution of a Thermostable D-Tagatose Epimerase towards Improved Activity for Two Hexose Substrates.
Chembiochem, 16, 2015
|
|
7NNK
 
 | | Crystal structure of S116A mutant of hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1 in complex with hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, HpcH/HpaI aldolase, MAGNESIUM ION | | Authors: | Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-02-25 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
4PU5
 
 | | Shewanella oneidensis Toxin Antitoxin System Toxin Protein HipA Bound with AMPPNP and Mg | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Wen, Y, Behiels, E, Felix, J, Elegheert, J, Vergauwen, B, Devreese, B, Savvides, S. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Nucleic Acids Res., 42, 2014
|
|
4PU4
 
 | | Shewanella oneidensis MR-1 Toxin Antitoxin System HipA, HipB and its operator DNA complex (space group P21) | | Descriptor: | Operator DNA, Toxin-antitoxin system antidote transcriptional repressor Xre family, Toxin-antitoxin system toxin HipA family | | Authors: | Wen, Y, Behiels, E, Felix, J, Elegheert, J, Vergauwen, B, Devreese, B, Savvides, S. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.786 Å) | | Cite: | The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Nucleic Acids Res., 42, 2014
|
|
6YKP
 
 | |
4PU7
 
 | | Shewanella oneidensis Toxin Antitoxin System Antitoxin Protein HipB Resolution 1.85 | | Descriptor: | Toxin-antitoxin system antidote transcriptional repressor Xre family | | Authors: | Wen, Y, Behiels, E, Felix, J, Elegheert, J, Vergauwen, B, Devreese, B, Savvides, S. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Nucleic Acids Res., 42, 2014
|
|
2M5T
 
 | | Solution structure of the 2A proteinase from a common cold agent, human rhinovirus RV-C02, strain W12 | | Descriptor: | ZINC ION, human rhinovirus 2A proteinase | | Authors: | Lee, W, Frederick, R, Tonelli, M, Troupis, A.T, Reinin, N, Suchy, F.P, Moyer, K, Watters, K, Aceti, D, Palmenberg, A.C, Markley, J.L. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the 2A Protease from a Common Cold Agent, Human Rhinovirus C2, Strain W12.
Plos One, 9, 2014
|
|
7O7G
 
 | | Crystal structure of the Shewanella oneidensis MR1 MtrC mutant H561M | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Edwards, M.J, van Wonderen, J.H, Newton-Payne, S.E, Butt, J.N, Clarke, T.A. | | Deposit date: | 2021-04-13 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nanosecond heme-to-heme electron transfer rates in a multiheme cytochrome nanowire reported by a spectrally unique His/Met-ligated heme.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YKM
 
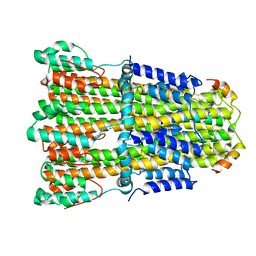 | | Structure of C. jejuni MotAB | | Descriptor: | Chemotaxis protein MotA, putative, Chemotaxis protein MotB | | Authors: | Santiveri, M, Roa-Eguiara, A, Taylor, N.M.I. | | Deposit date: | 2020-04-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and Function of Stator Units of the Bacterial Flagellar Motor.
Cell, 183, 2020
|
|
6Y3D
 
 | | X-ray structure of thermophilic C-phycocyanin from Galdiera phlegrea | | Descriptor: | ACETATE ION, C-phycocyanin alpha chain, C-phycocyanin beta chain, ... | | Authors: | Ferraro, G, Lucignano, R, Marseglia, A, Merlino, A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of C-phycocyanin from Galdieria phlegrea: Determinants of thermostability and comparison with a C-phycocyanin in the entire phycobilisome.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
8ADQ
 
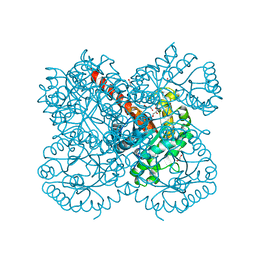 | | Crystal structure of holo-SwHPA-Mg (hydroxy ketone aldolase) from Sphingomonas wittichii RW1 in complex with hydroxypyruvate and D-Glyceraldehyde | | Descriptor: | 3-HYDROXYPYRUVIC ACID, BROMIDE ION, D-Glyceraldehyde, ... | | Authors: | Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2022-07-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6EZF
 
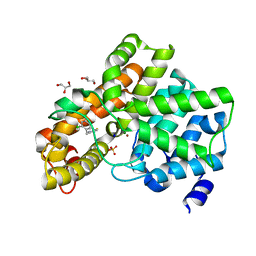 | | PDE2 in complex with molecule 5 | | Descriptor: | 6-[(2,4-dichlorophenyl)methyl]pyridazine-3-thiol, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tresadern, G, Perez-Benito, L, Keraenen, H, van Vlijmen, H. | | Deposit date: | 2017-11-15 | | Release date: | 2018-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Predicting Binding Free Energies of PDE2 Inhibitors. The Difficulties of Protein Conformation.
Sci Rep, 8, 2018
|
|
7NUJ
 
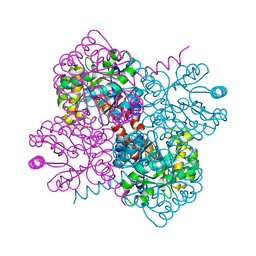 | | Crystal structure of holo-SwHPA-Mg (hydroxy ketone aldolase) from Sphingomonas wittichii RW1 | | Descriptor: | HpcH/HpaI aldolase, MAGNESIUM ION | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-03-12 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7O5R
 
 | | Crystal structure of holo-SwHPA-Mn (hydroxyketoacid aldolase) from Sphingomonas wittichii RW1 | | Descriptor: | BROMIDE ION, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-09 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7O9R
 
 | | Crystal structure of holo-H44A mutant of Hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1 | | Descriptor: | BROMIDE ION, DI(HYDROXYETHYL)ETHER, HpcH/HpaI aldolase, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-16 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7O87
 
 | | Crystal structure of holo-F210W mutant of Hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1 in complex with hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, BROMIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-14 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7O5W
 
 | | Crystal structure of holo-F210W mutant of Hydroxy ketone aldolase (SwHKA)from Sphingomonas wittichii RW1 | | Descriptor: | BROMIDE ION, DI(HYDROXYETHYL)ETHER, HpcH/HpaI aldolase, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-09 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7OBU
 
 | | Crystal structure of holo-F210W mutant of Hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1, with the active site in the resting and the active state | | Descriptor: | 3-HYDROXYPYRUVIC ACID, HpcH/HpaI aldolase, MAGNESIUM ION, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-23 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7O5I
 
 | | Crystal structure of apo-SwHKA (Hydroxy ketone aldolase) from Sphingomonas wittichii RW1 | | Descriptor: | BROMIDE ION, DI(HYDROXYETHYL)ETHER, HpcH/HpaI aldolase, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7O5V
 
 | | Crystal structure of holo-H44A mutant of Hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1, in complex with Hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, BROMIDE ION, HpcH/HpaI aldolase, ... | | Authors: | Laustsen, J, Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-04-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8B30
 
 | |
