2MDR
 
 | | Solution structure of the third double-stranded RNA-binding domain (dsRBD3) of human adenosine-deaminase ADAR1 | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Barraud, P, Banerjee, S, Mohamed, W.I, Jantsch, M.F, Allain, F.H. | | Deposit date: | 2013-09-17 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A bimodular nuclear localization signal assembled via an extended double-stranded RNA-binding domain acts as an RNA-sensing signal for transportin 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MDT
 
 | |
2MDU
 
 | | Circular Permutant of the WW Domain with Loop 1 Excised | | Descriptor: | Pin1 WW domain | | Authors: | Kier, B.L. | | Deposit date: | 2013-09-18 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Circular Permutation of a WW Domain: Folding Still Occurs after Excising the Turn of the Folding-Nucleating Hairpin.
J.Am.Chem.Soc., 136, 2014
|
|
2MDV
 
 | |
2MDW
 
 | |
2MDX
 
 | |
2MDZ
 
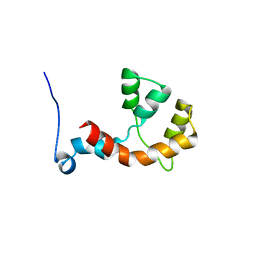 | | NMR structure of the Paracoccus denitrificans Z-subunit determined in the presence of ADP | | Descriptor: | Uncharacterized protein | | Authors: | Serrano, P, Geralt, M, Wuthrich, K, Morales-Rios, E, Zarco-Zavala, M, Garcia-Trejo, J.J, Dutta, S.K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the putative ATPase regulatory protein YP_916642.1 from Paracoccus denitrificans
To be Published
|
|
2ME0
 
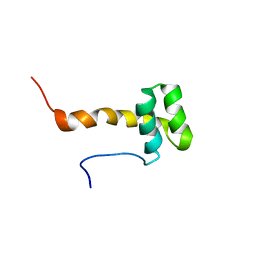 | |
2ME1
 
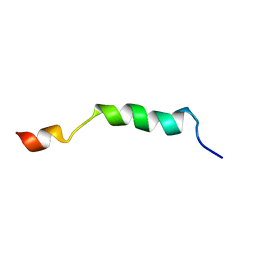 | | HIV-1 gp41 clade B double alanine mutant Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Gp41 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
2ME2
 
 | | HIV-1 gp41 clade C Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
2ME3
 
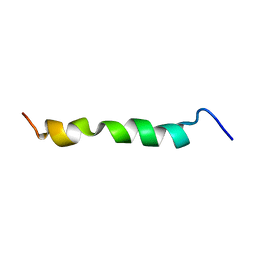 | | HIV-1 gp41 clade C Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
2ME4
 
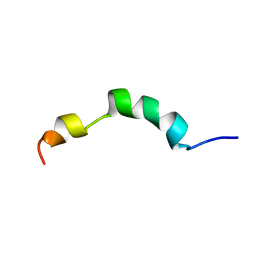 | | HIV-1 gp41 clade C Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
2ME6
 
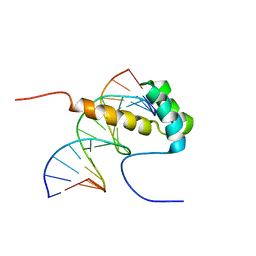 | | NMR Structure of the homeodomain transcription factor Gbx1 from Homo sapiens in complex with the DNA sequence CGACTAATTAGTCG | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*TP*AP*AP*TP*TP*AP*GP*TP*CP*G)-3'), Homeobox protein GBX-1 | | Authors: | Proudfoot, A, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the homeodomain transcription factor Gbx1 from Homo sapiens in complex with the DNA sequence CGACTAATTAGTCG
To be Published
|
|
2ME7
 
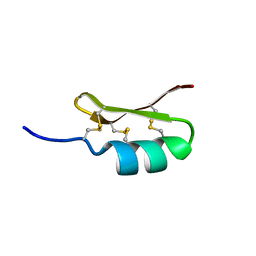 | |
2ME8
 
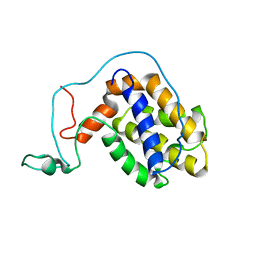 | |
2ME9
 
 | |
2MEA
 
 | |
2MEB
 
 | | CHANGES IN CONFORMATIONAL STABILITY OF A SERIES OF MUTANT HUMAN LYSOZYMES AT CONSTANT POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
2MEC
 
 | |
2MED
 
 | | CONTRIBUTION OF HYDROPHOBIC EFFECT TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
2MEE
 
 | |
2MEF
 
 | | CONTRIBUTION OF HYDROPHOBIC EFFECT TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-04 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
2MEG
 
 | | CHANGES IN CONFORMATIONAL STABILITY OF A SERIES OF MUTANT HUMAN LYSOZYMES AT CONSTANT POSITIONS. | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
2MEH
 
 | | CONTRIBUTION OF HYDROPHOBIC EFFECT TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
2MEI
 
 | | CONTRIBUTION OF HYDROPHOBIC EFFECT TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-02 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
