1TUL
 
 | | STRUCTURE OF TLP20 | | Descriptor: | TLP20 | | Authors: | Rayment, I, Holden, H.M. | | Deposit date: | 1996-08-17 | | Release date: | 1997-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular structure of a proteolytic fragment of TLP20.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
297D
 
 | |
1PUP
 
 | |
1WAV
 
 | | CRYSTAL STRUCTURE OF FORM B MONOCLINIC CRYSTAL OF INSULIN | | Descriptor: | INSULIN, PHENOL, ZINC ION | | Authors: | Liang, D.-C, Ding, J.-H, Chang, W.-R, Wan, Z.-L. | | Deposit date: | 1996-02-28 | | Release date: | 1997-02-28 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular replacement study on form-B monoclinic crystal of insulin.
Sci.China, Ser.C: Life Sci., 39, 1996
|
|
1PMI
 
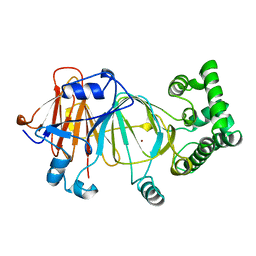 | | Candida Albicans Phosphomannose Isomerase | | Descriptor: | PHOSPHOMANNOSE ISOMERASE, ZINC ION | | Authors: | Cleasby, A, Skarzynski, T, Wonacott, A, Davies, G.J, Hubbard, R.E, Proudfoot, A.E.I, Wells, T.N.C, Payton, M.A, Bernard, A.R. | | Deposit date: | 1996-04-03 | | Release date: | 1997-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The x-ray crystal structure of phosphomannose isomerase from Candida albicans at 1.7 angstrom resolution.
Nat.Struct.Biol., 3, 1996
|
|
1GIF
 
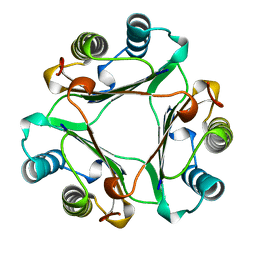 | | HUMAN GLYCOSYLATION-INHIBITING FACTOR | | Descriptor: | GLYCOSYLATION-INHIBITING FACTOR | | Authors: | Kato, Y, Kuroki, R. | | Deposit date: | 1996-02-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human glycosylation-inhibiting factor is a trimeric barrel with three 6-stranded beta-sheets.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1HPK
 
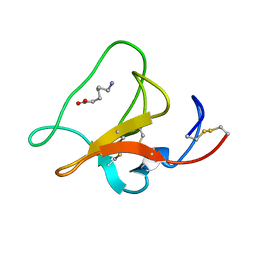 | |
1GOT
 
 | | HETEROTRIMERIC COMPLEX OF A GT-ALPHA/GI-ALPHA CHIMERA AND THE GT-BETA-GAMMA SUBUNITS | | Descriptor: | GT-ALPHA/GI-ALPHA CHIMERA, GT-BETA, GT-GAMMA, ... | | Authors: | Lambright, D.G, Sondek, J, Bohm, A, Skiba, N.P, Hamm, H.E, Sigler, P.B. | | Deposit date: | 1996-08-07 | | Release date: | 1997-03-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of a heterotrimeric G protein.
Nature, 379, 1996
|
|
3BTO
 
 | |
1ZWD
 
 | |
1SPG
 
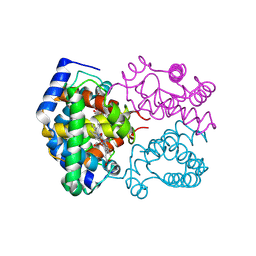 | |
1GZB
 
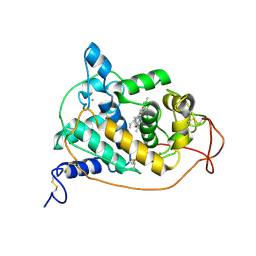 | | PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PEROXIDASE, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pentacoordination of the heme iron of Arthromyces ramosus peroxidase shown by a 1.8 A resolution crystallographic study at pH 4.5.
FEBS Lett., 378, 1996
|
|
1GZA
 
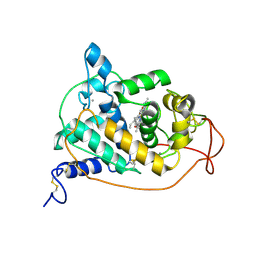 | | PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Binding of iodide to Arthromyces ramosus peroxidase investigated with X-ray crystallographic analysis, 1H and 127I NMR spectroscopy, and steady-state kinetics.
J.Biol.Chem., 272, 1997
|
|
1IYV
 
 | |
1SPU
 
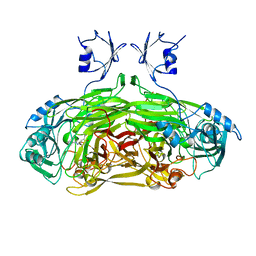 | | STRUCTURE OF OXIDOREDUCTASE | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Wilmot, C.M, Phillips, S.E.V. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic mechanism of the quinoenzyme amine oxidase from Escherichia coli: exploring the reductive half-reaction.
Biochemistry, 36, 1997
|
|
2LFB
 
 | | HOMEODOMAIN FROM RAT LIVER LFB1/HNF1 TRANSCRIPTION FACTOR, NMR, 20 STRUCTURES | | Descriptor: | LFB1/HNF1 TRANSCRIPTION FACTOR | | Authors: | Schott, O, Billeter, M, Leiting, B, Wider, G, Wuthrich, K. | | Deposit date: | 1996-12-12 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the non-classical homeodomain from the rat liver LFB1/HNF1 transcription factor.
J.Mol.Biol., 267, 1997
|
|
1DJA
 
 | |
1IYU
 
 | |
3CEL
 
 | | ACTIVE-SITE MUTANT E212Q DETERMINED AT PH 6.0 WITH CELLOBIOSE BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
1WBR
 
 | |
1HCW
 
 | |
1TFB
 
 | |
1TBD
 
 | | SOLUTION STRUCTURE OF THE ORIGIN DNA BINDING DOMAIN OF SV40 T-ANTIGEN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SV40 T-ANTIGEN | | Authors: | Luo, X, Sanford, D.G, Bullock, P.A, Bachovchin, W.W. | | Deposit date: | 1996-11-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the origin DNA-binding domain of SV40 T-antigen.
Nat.Struct.Biol., 3, 1996
|
|
1AII
 
 | | ANNEXIN III | | Descriptor: | ANNEXIN III, CALCIUM ION, ETHANOLAMINE, ... | | Authors: | Lewit-Bentley, A, Perron, B. | | Deposit date: | 1996-11-28 | | Release date: | 1997-03-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Can enzymatic activity, or otherwise, be inferred from structural studies of annexin III?
J.Biol.Chem., 272, 1997
|
|
1VZB
 
 | |
