1X4A
 
 | | Solution structure of RRM domain in splicing factor SF2 | | Descriptor: | splicing factor, arginine/serine-rich 1 (splicing factor 2, alternate splicing factor) variant | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in splicing factor SF2
To be Published
|
|
1X4Q
 
 | | Solution structure of PWI domain in U4/U6 small nuclear ribonucleoprotein Prp3(hPrp3) | | Descriptor: | U4/U6 small nuclear ribonucleoprotein Prp3 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PWI domain in U4/U6 small nuclear ribonucleoprotein Prp3(hPrp3)
To be Published
|
|
1WGQ
 
 | | Solution Structure of the Pleckstrin Homology Domain of Mouse Ethanol Decreased 4 Protein | | Descriptor: | FYVE, RhoGEF and PH domain containing 6; ethanol decreased 4 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pleckstrin Homology Domain of Mouse Ethanol Decreased 4 Protein
To be Published
|
|
1WHA
 
 | | Solution structure of the second PDZ domain of human scribble (KIAA0147 protein). | | Descriptor: | KIAA0147 protein | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second PDZ domain of human scribble (KIAA0147 protein).
To be Published
|
|
1WHY
 
 | | Solution structure of the RNA recognition motif from hypothetical RNA binding protein BC052180 | | Descriptor: | hypothetical protein RIKEN cDNA 1810017N16 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA recognition motif from hypothetical RNA binding protein BC052180
To be Published
|
|
1WHX
 
 | | Solution structure of the second RNA binding domain from hypothetical protein BAB23448 | | Descriptor: | hypothetical protein RIKEN CDNA 1200009A02 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RNA binding domain from hypothetical protein BAB23448
To be Published
|
|
1X4R
 
 | | Solution structure of WWE domain in Parp14 protein | | Descriptor: | Parp14 protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of WWE domain in Parp14 protein
To be Published
|
|
1X5S
 
 | | Solution structure of RRM domain in A18 hnRNP | | Descriptor: | Cold-inducible RNA-binding protein | | Authors: | Sato, A, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-16 | | Release date: | 2005-11-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in A18 hnRNP
To be Published
|
|
1WWZ
 
 | |
1WY9
 
 | | Crystal structure of microglia-specific protein, Iba1 | | Descriptor: | Allograft inflammatory factor 1, CALCIUM ION | | Authors: | Yamada, M, Imai, Y, Kohsaka, S, Kamitori, S. | | Deposit date: | 2005-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structures of the Microglia/Macrophage-specific Protein Iba1 from Human and Mouse Demonstrate Novel Molecular Conformation Change Induced by Calcium binding
J.Mol.Biol., 364, 2006
|
|
1X49
 
 | | Solution structure of the first DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first DSRM domain in Interferon-induced, double-stranded RNA-activated protein kinase
To be Published
|
|
1X4N
 
 | | Solution structure of KH domain in FUSE binding protein 1 | | Descriptor: | Far upstream element binding protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of KH domain in FUSE binding protein 1
To be Published
|
|
1X2I
 
 | | Crystal Structure Of Archaeal Xpf/Mus81 Homolog, Hef From Pyrococcus Furiosus, Helix-hairpin-helix Domain | | Descriptor: | Hef helicase/nuclease | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2005-04-24 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Analyses of an Archaeal XPF/Rad1/Mus81 Nuclease: Asymmetric DNA Binding and Cleavage Mechanisms
STRUCTURE, 13, 2005
|
|
1X47
 
 | | Solution structure of DSRM domain in DGCR8 protein | | Descriptor: | DGCR8 protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of DSRM domain in DGCR8 protein
To be Published
|
|
1X4P
 
 | | Solution structure of SURP domain in SFRS14 protei | | Descriptor: | Putative splicing factor, arginine/serine-rich 14 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SURP domain in SFRS14 protei
To be Published
|
|
1JO3
 
 | |
1JRV
 
 | | SOLUTION STRUCTURE OF DAATAA DNA BULGE | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*CP*GP*AP*AP*TP*AP*AP*GP*CP*TP*AP*CP*G)-3' | | Authors: | Gollmick, F.A, Lorenz, M, Dornberger, U, von Langen, J, Diekmann, S, Fritzsche, H. | | Deposit date: | 2001-08-15 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dAATAA and dAAUAA DNA bulges.
Nucleic Acids Res., 30, 2002
|
|
1X8T
 
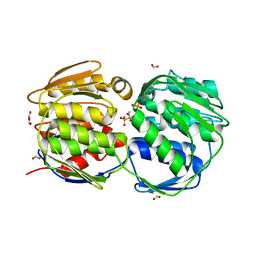 | | EPSPS liganded with the (R)-phosphonate analog of the tetrahedral reaction intermediate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, [3R-[3A,4A,5B(R*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID | | Authors: | Priestman, M.A, Healy, M.L, Becker, A, Alberg, D.G, Bartlett, P.A, Schonbrunn, E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction of phosphonate analogues of the tetrahedral reaction intermediate with 5-enolpyruvylshikimate-3-phosphate synthase in atomic detail.
Biochemistry, 44, 2005
|
|
1X75
 
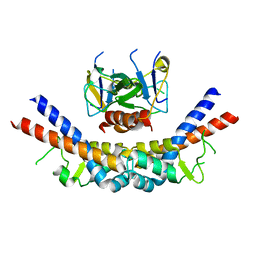 | | CcdB:GyrA14 complex | | Descriptor: | Cytotoxic protein ccdB, DNA gyrase subunit A | | Authors: | Dao-Thi, M.H, Van Melderen, L, De Genst, E, Wyns, L, Loris, R. | | Deposit date: | 2004-08-13 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Basis of Gyrase Poisoning by the Addiction Toxin CcdB
J.Mol.Biol., 348, 2005
|
|
1XCS
 
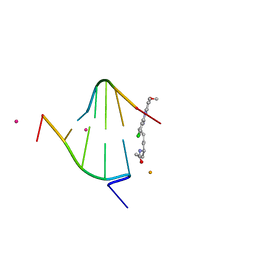 | | structure of oligonucleotide/drug complex | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9-[(5-(ACETYLAMINO)-6-{[(1S,4R)-8-AMINO-4-[((2R)-6-AMINO-2-{2-[(1S)-5-AMINO-1-FORMYLPENTYL]HYDRAZINO}HEXANOYL)AMINO]-1-(4-AMINOBUTYL)-2,3-DIOXOOCTYL]AMINO}-6-OXOHEXYL)AMINO]-6-CHLORO-2-METHOXYACRIDINIUM, BARIUM ION, ... | | Authors: | Valls, N, Steiner, R.A, Wright, G, Murshudov, G.N, Subirana, J.A. | | Deposit date: | 2004-09-03 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Variable role of ions in two drug intercalation complexes of DNA
J.Biol.Inorg.Chem., 10, 2005
|
|
1JUE
 
 | | 1.8 A resolution structure of native lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | ACETIC ACID, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-24 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JRW
 
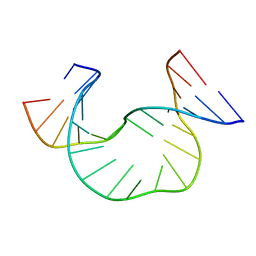 | | Solution Structure of dAATAA DNA Bulge | | Descriptor: | 5'-D(*CP*GP*TP*AP*GP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*CP*GP*AP*AP*TP*AP*AP*GP*CP*TP*AP*CP*G)-3' | | Authors: | Gollmick, F.A, Lorenz, M, Dornberger, U, von Langen, J, Diekmann, S, Fritzsche, H. | | Deposit date: | 2001-08-15 | | Release date: | 2002-08-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dAATAA and dAAUAA DNA bulges.
Nucleic Acids Res., 30, 2002
|
|
1JSI
 
 | | CRYSTAL STRUCTURE OF H9 HAEMAGGLUTININ BOUND TO LSTC RECEPTOR ANALOG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ha, Y, Stevens, D.J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 2001-08-17 | | Release date: | 2001-09-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JVY
 
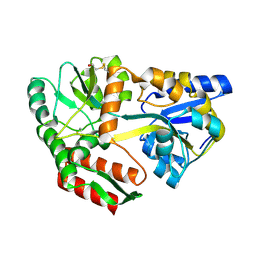 | | Maltodextrin-binding protein variant D207C/A301GS/P316C with beta-mercaptoethanol mixed disulfides | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltodextrin-binding protein | | Authors: | Srinivasan, U, Iyer, G.H, Przybycien, T.A, Samsonoff, W.A, Bell, J.A. | | Deposit date: | 2001-08-31 | | Release date: | 2001-09-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystine: Fibrous Biomolecular Material from Protein Crystals Cross-linked in a Specific Geometry
Protein Eng., 15, 2002
|
|
1X8J
 
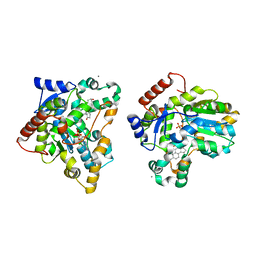 | | Crystal structure of retinol dehydratase in complex with androsterone and inactive cofactor PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Androsterone, CALCIUM ION, ... | | Authors: | Pakhomova, S, Buck, J, Newcomer, M.E. | | Deposit date: | 2004-08-18 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structures of the unique sulfotransferase retinol dehydratase with product and inhibitors provide insight into enzyme mechanism and inhibition.
Protein Sci., 14, 2005
|
|
