3IKI
 
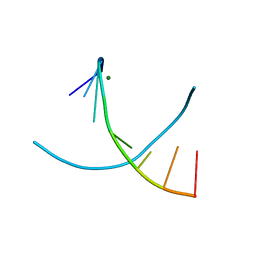 | | 5-SMe-dU containing DNA octamer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(US2)P*AP*CP*AP*C)-3', MAGNESIUM ION | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | Deposit date: | 2009-08-05 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Hydrogen bond formation between the naturally modified nucleobase and phosphate backbone.
Nucleic Acids Res., 40, 2012
|
|
3I8X
 
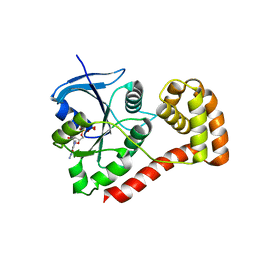 | | Structure of the cytosolic domain of E. coli FeoB, GDP-bound form | | Descriptor: | Ferrous iron transport protein B, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Petermann, N, Hansen, G, Hogg, T, Hilgenfeld, R. | | Deposit date: | 2009-07-10 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the intrinsic GTPase and GDI activities of FeoB, a prokaryotic transmembrane GTP/GDP-binding protein
To be Published
|
|
2R3N
 
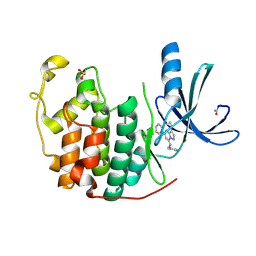 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 3-cyclopropyl-5-phenyl-N-(pyridin-3-ylmethyl)pyrazolo[1,5-a]pyrimidin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
2R3G
 
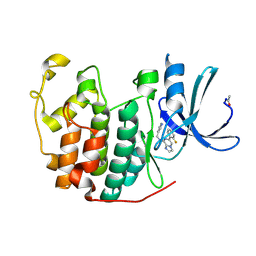 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 6-(2-fluorophenyl)-N-(pyridin-3-ylmethyl)imidazo[1,2-a]pyrazin-8-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
3C41
 
 | | ABC protein ArtP in complex with AMP-PNP/Mg2+ | | Descriptor: | Amino acid ABC transporter (ArtP), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Thaben, P.F, Eckey, V, Scheffel, F, Saenger, W, Schneider, E, Vahedi-Faridi, A. | | Deposit date: | 2008-01-29 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures of the ATP-binding cassette (ABC) Protein ArtP from Geobacillus stearothermophilus in complexes with nucleotides and nucleotide analogs reveal an intermediate semiclosed dimer in the post hydrolyses state and an asymmetry in the dimerisation region
To be Published
|
|
2R44
 
 | |
3IU5
 
 | | Crystal structure of the first bromodomain of human poly-bromodomain containing protein 1 (PB1) | | Descriptor: | Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-30 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
2QFS
 
 | | E.coli EPSP synthase Pro101Ser liganded with S3P | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E, Healy-Fried, M.L. | | Deposit date: | 2007-06-28 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis of glyphosate tolerance resulting from mutations of Pro101 in Escherichia coli 5-enolpyruvylshikimate-3-phosphate synthase.
J.Biol.Chem., 282, 2007
|
|
3C7A
 
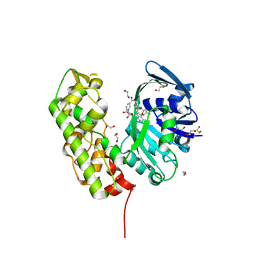 | | A structural basis for substrate and stereo selectivity in octopine dehydrogenase (ODH-NADH) | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Octopine dehydrogenase | | Authors: | Smits, S.H.J, Mueller, A, Schmitt, L, Grieshaber, M.K. | | Deposit date: | 2008-02-07 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for substrate selectivity and stereoselectivity in octopine dehydrogenase from Pecten maximus.
J.Mol.Biol., 381, 2008
|
|
2QIQ
 
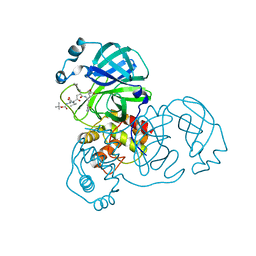 | |
2QJ9
 
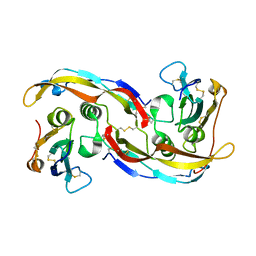 | | Crystal structure analysis of BMP-2 in complex with BMPR-IA variant B1 | | Descriptor: | Bone morphogenetic protein 2, Bone morphogenetic protein receptor type IA | | Authors: | Kotzsch, A, Mueller, T.D. | | Deposit date: | 2007-07-06 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure analysis of bone morphogenetic protein-2 type I receptor complexes reveals a mechanism of receptor inactivation in juvenile polyposis syndrome.
J.Biol.Chem., 283, 2008
|
|
3IF6
 
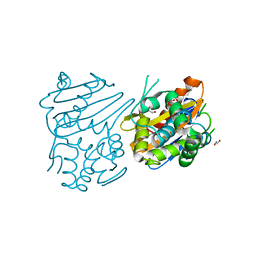 | | Crystal structure of OXA-46 beta-lactamase from P. aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, L(+)-TARTARIC ACID, ... | | Authors: | Docquier, J.D, Benvenuti, M, Calderone, V, Giuliani, F, Kapetis, D, De Luca, F, Rossolini, G.M, Mangani, S. | | Deposit date: | 2009-07-24 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the narrow-spectrum OXA-46 class D beta-lactamase: relationship between active-site lysine carbamylation and inhibition by polycarboxylates
Antimicrob.Agents Chemother., 54, 2010
|
|
3IFS
 
 | | 2.0 Angstrom Resolution Crystal Structure of Glucose-6-phosphate Isomerase (pgi) from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, Glucose-6-phosphate isomerase, LITHIUM ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Onopriyenko, O, Gordon, E, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-24 | | Release date: | 2009-08-11 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of Glucose-6-phosphate Isomerase (pgi) from Bacillus anthracis.
To be Published
|
|
3IHD
 
 | |
3BK5
 
 | |
3C04
 
 | |
2QJA
 
 | | Crystal structure analysis of BMP-2 in complex with BMPR-IA variant B12 | | Descriptor: | Bone morphogenetic protein 2, Bone morphogenetic protein receptor type IA | | Authors: | Kotzsch, A, Mueller, T.D. | | Deposit date: | 2007-07-06 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure analysis of bone morphogenetic protein-2 type I receptor complexes reveals a mechanism of receptor inactivation in juvenile polyposis syndrome.
J.Biol.Chem., 283, 2008
|
|
2QK7
 
 | | A covalent S-F heterodimer of staphylococcal gamma-hemolysin | | Descriptor: | Gamma-hemolysin component A, Gamma-hemolysin component B | | Authors: | Roblin, P, Guillet, V, Maveyraud, L, Mourey, L. | | Deposit date: | 2007-07-10 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A covalent S-F heterodimer of leucotoxin reveals molecular plasticity of beta-barrel pore-forming toxins.
Proteins, 71, 2008
|
|
3C11
 
 | |
3BQE
 
 | |
3D1J
 
 | |
2QTJ
 
 | | Solution structure of human dimeric immunoglobulin A | | Descriptor: | Ig alpha-1 chain C region, Kappa light chain IgA1 | | Authors: | Bonner, A, Furtado, P.B, Almogren, A, Kerr, M.A, Perkins, S.J. | | Deposit date: | 2007-08-02 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Implications of the near-planar solution structure of human myeloma dimeric IgA1 for mucosal immunity and IgA nephropathy
J.Immunol., 180, 2008
|
|
2QSS
 
 | | Bovine hemoglobin at pH 6.3 | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Aranda IV, R, Richards, M.P, Phillips Jr, G.N. | | Deposit date: | 2007-07-31 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of fish versus mammalian hemoglobins: Effect of the heme pocket environment on autooxidation and hemin loss.
Proteins, 75, 2008
|
|
3I79
 
 | |
3D53
 
 | |
