4LK9
 
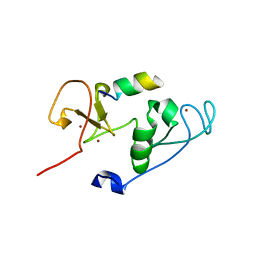 | | Crystal Structure of MOZ double PHD finger histone H3 tail complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-07 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
6AML
 
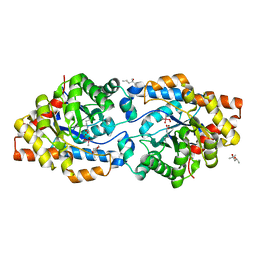 | | Phosphotriesterase variant S8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Phosphotriesterase variant S8
To Be Published
|
|
1UVO
 
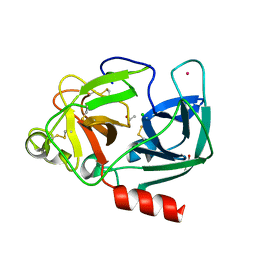 | | Structure Of The Complex Of Porcine Pancreatic Elastase In Complex With Cadmium Refined At 1.85 A Resolution (Crystal A) | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Weiss, M.S, Panjikar, S, Mueller-Dieckmann, C, Tucker, P.A. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | On the Influence of the Incident Photon Energy on the Radiation Damage in Crystalline Biological Samples
J.Synchrotron Radiat., 12, 2005
|
|
5YVS
 
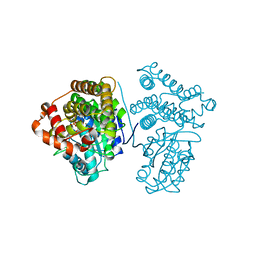 | | Crystal Structure of the archaeal halo-thermophilic Red Sea brine pool alcohol dehydrogenase ADH/D1 bound to NADP | | Descriptor: | MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, alcohol dehydrogenase | | Authors: | Groetzinger, S.W, Strillinger, E, Frank, A, Eppinger, J, Groll, M, Arold, S.T. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes
ACS Chem. Biol., 13, 2018
|
|
1XPM
 
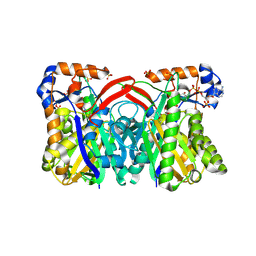 | | Crystal Structure of Staphylococcus aureus HMG-COA Synthase with HMG-CoA and Acetoacetyl-COA and Acetylated Cysteine | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, 3-hydroxy-3-methylglutaryl CoA synthase, ACETOACETYL-COENZYME A, ... | | Authors: | Theisen, M.J, Misra, I, Saadat, D, Campobasso, N, Miziorko, H.M, Harrison, D.H.T. | | Deposit date: | 2004-10-08 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 3-hydroxy-3-methylglutaryl-CoA synthase intermediate complex observed in "real-time"
Proc.Natl.Acad.Sci.USA, 47, 2004
|
|
3TEN
 
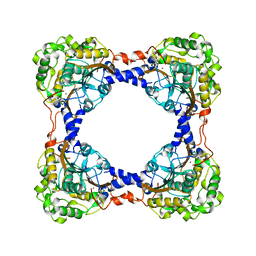 | | Holo form of carbon disulfide hydrolase | | Descriptor: | CS2 hydrolase, ZINC ION | | Authors: | Smeulders, M.J, Barends, T.R.M.B, Pol, A, Scherer, A, Zandvoort, M.H, Udvarhelyi, A, Khadem, A, Menzel, A, Hermans, J, Shoeman, R.L, Wessels, H.J.C.T, van den Heuvel, L.P, Russ, L, Schlichting, I, Jetten, M.S.M, Op den Camp, H.J.M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolution of a new enzyme for carbon disulphide conversion by an acidothermophilic archaeon.
Nature, 478, 2011
|
|
1XQL
 
 | | Effect of a Y265F Mutant on the Transamination Based Cycloserine Inactivation of Alanine Racemase | | Descriptor: | (5-HYDROXY-4-{[(3-HYDROXYISOXAZOL-4-YL)AMINO]METHYL}-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, (R)-4-AMINO-ISOXAZOLIDIN-3-ONE, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Fenn, T.D, Holyoak, T, Stamper, G.F, Ringe, D. | | Deposit date: | 2004-10-12 | | Release date: | 2005-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of a Y265F Mutant on the Transamination-Based Cycloserine Inactivation of Alanine Racemase
Biochemistry, 44, 2005
|
|
5YVR
 
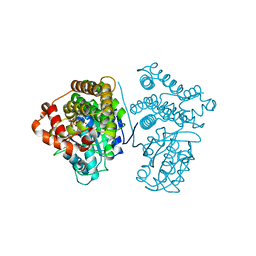 | | Crystal Structure of the H277A mutant of ADH/D1, an archaeal halo-thermophilic Red Sea brine pool alcohol dehydrogenase | | Descriptor: | MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, alcohol dehydrogenase | | Authors: | Groetzinger, S.W, Strillinger, E, Frank, A, Eppinger, J, Groll, M, Arold, S.T. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes
ACS Chem. Biol., 13, 2018
|
|
5YVM
 
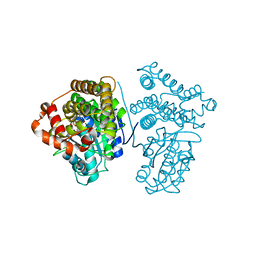 | | Crystal Structure of the archaeal halo-thermophilic Red Sea brine pool alcohol dehydrogenase ADH/D1 bound to NZQ | | Descriptor: | 5,6-DIHYDROXY-NADP, MANGANESE (II) ION, alcohol dehydrogenase | | Authors: | Groetzinger, S.W, Strillinger, E, Frank, A, Eppinger, J, Groll, M, Arold, S.T. | | Deposit date: | 2017-11-26 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes
ACS Chem. Biol., 13, 2018
|
|
1YHK
 
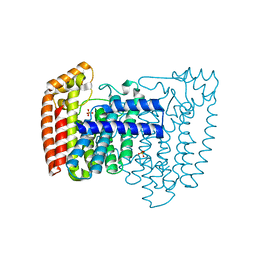 | | Trypanosoma cruzi farnesyl diphosphate synthase | | Descriptor: | SULFATE ION, farnesyl pyrophosphate synthase | | Authors: | Gabelli, S.B, McLellan, J.S, Montalvetti, A, Oldfield, E, Docampo, R, Amzel, L.M. | | Deposit date: | 2005-01-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: Implications for drug design.
Proteins, 62, 2005
|
|
4MAE
 
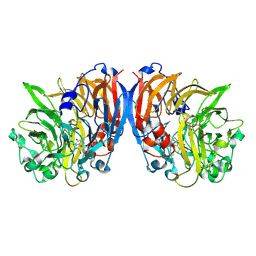 | | Methanol dehydrogenase from Methylacidiphilum fumariolicum SolV | | Descriptor: | CERIUM (III) ION, Methanol dehydrogenase, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Pol, A.J, Barends, T.R.M, Dietl, A, Khadem, A.F, Eygenstein, J, Jetten, M.S.M, Op den Camp, H.J.M. | | Deposit date: | 2013-08-16 | | Release date: | 2013-09-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare earth metals are essential for methanotrophic life in volcanic mudpots.
ENVIRON.MICROBIOL., 16, 2014
|
|
2A79
 
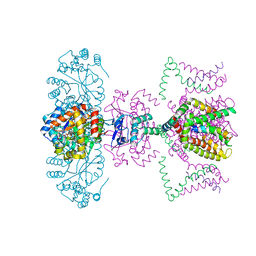 | | Mammalian Shaker Kv1.2 potassium channel- beta subunit complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2, ... | | Authors: | Long, S.B, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a mammalian voltage-dependent Shaker family K+ channel.
Science, 309, 2005
|
|
1Y19
 
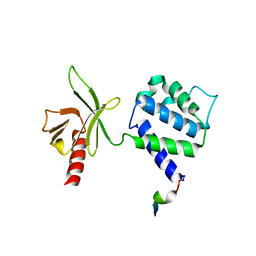 | | Structural basis for phosphatidylinositol phosphate kinase type I-gamma binding to talin at focal adhesions | | Descriptor: | Phosphatidylinositol-4-phosphate 5-kinase, type 1 gamma, Talin 1 | | Authors: | de Pereda, J.M, Wegener, K, Santelli, E, Bate, N, Ginsberg, M.H, Critchley, D.R, Campbell, I.D, Liddington, R.C. | | Deposit date: | 2004-11-17 | | Release date: | 2005-01-04 | | Last modified: | 2016-11-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural bases for phosphatidylinositol phosphate kinase type I-gamma binding to talin at focal adhesions
J.Biol.Chem., 280, 2005
|
|
3A3I
 
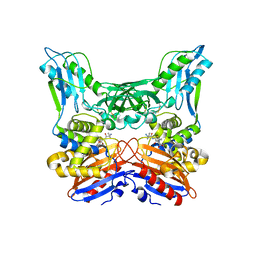 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae, complexed with ampicillin (AIX) | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4 | | Authors: | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2009-06-12 | | Release date: | 2009-12-22 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
1YHL
 
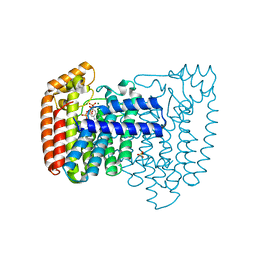 | | Structure of the complex of Trypanosoma cruzi farnesyl diphosphate synthase with risedronate, dmapp and mg+2 | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, DIMETHYLALLYL DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gabelli, S.B, McLellan, J.S, Montalvetti, A, Oldfield, E, Docampo, R, Amzel, L.M. | | Deposit date: | 2005-01-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: Implications for drug design.
Proteins, 62, 2005
|
|
5IGA
 
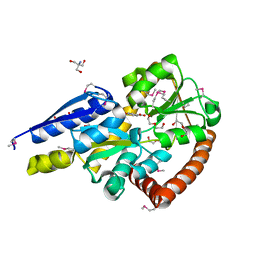 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with co-purified parahydroxybenzoate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, P-HYDROXYBENZOIC ACID, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-02-27 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with co-purified parahydroxybenzoate
To be published
|
|
3WNR
 
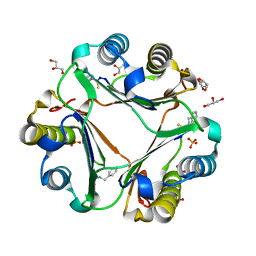 | | Multiple binding modes of benzyl isothiocyanate inhibitor complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | CHLORIDE ION, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Spencer, E.S, Dale, E.J, Gommans, A.L, Rutledge, M.T, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Multiple binding modes of isothiocyanates that inhibit macrophage migration inhibitory factor
Eur.J.Med.Chem., 93, 2015
|
|
1YHM
 
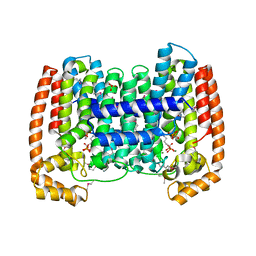 | | Structure of the complex of Trypanosoma cruzi farnesyl disphosphate synthase with alendronate, Isopentenyl diphosphate and mg+2 | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, MAGNESIUM ION, ... | | Authors: | Gabelli, S.B, McLellan, J.S, Montalvetti, A, Oldfield, E, Docampo, R, Amzel, L.M. | | Deposit date: | 2005-01-09 | | Release date: | 2005-12-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: Implications for drug design.
Proteins, 62, 2005
|
|
1YIO
 
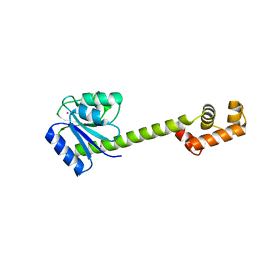 | | Crystallographic structure of response regulator StyR from Pseudomonas fluorescens | | Descriptor: | MAGNESIUM ION, MERCURY (II) ION, response regulatory protein | | Authors: | Milani, M, Leoni, L, Rampioni, G, Zennaro, E, Ascenzi, P, Bolognesi, M. | | Deposit date: | 2005-01-12 | | Release date: | 2005-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Active-like Structure in the Unphosphorylated StyR Response Regulator Suggests a Phosphorylation- Dependent Allosteric Activation Mechanism
STRUCTURE, 13, 2005
|
|
5IRB
 
 | | Structural insight into host cell surface retention of a 1.5-MDa bacterial ice-binding adhesin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Guo, S, Phippen, S, Campbell, R, Davies, P. | | Deposit date: | 2016-03-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5I7I
 
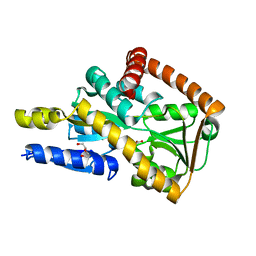 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-crystallized 3-hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, DI(HYDROXYETHYL)ETHER, TRAP solute Binding Protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-02-17 | | Release date: | 2017-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-purified 4-hydroxybenzoate
To Be Published
|
|
1DFN
 
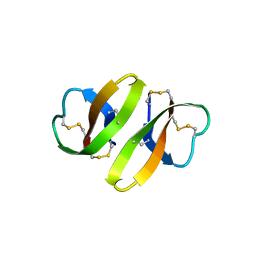 | | CRYSTAL STRUCTURE OF DEFENSIN HNP-3, AN AMPHIPHILIC DIMER: MECHANISMS OF MEMBRANE PERMEABILIZATION | | Descriptor: | DEFENSIN HNP-3 | | Authors: | Hill, C.P, Yee, J, Selsted, M.E, Eisenberg, D. | | Deposit date: | 1991-01-18 | | Release date: | 1992-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization.
Science, 251, 1991
|
|
1G5N
 
 | | ANNEXIN V COMPLEX WITH HEPARIN OLIGOSACCHARIDES | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ANNEXIN V, CALCIUM ION | | Authors: | Capila, I, Heraiz, M.J, Mo, Y.D, Mealy, T.R, Campos, B, Dedman, J.R, Linhardt, R.J, Seaton, B.A. | | Deposit date: | 2000-11-01 | | Release date: | 2001-06-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Annexin V--heparin oligosaccharide complex suggests heparan sulfate--mediated assembly on cell surfaces.
Structure, 9, 2001
|
|
4MV3
 
 | | Crystal Structure of Biotin Carboxylase from Haemophilus influenzae in Complex with AMPPCP and Bicarbonate | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, Biotin carboxylase, ... | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R.J, Waldrop, G.L. | | Deposit date: | 2013-09-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
2GVM
 
 | | Crystal structure of hydrophobin HFBI with detergent | | Descriptor: | Hydrophobin-1, LAURYL DIMETHYLAMINE-N-OXIDE, ZINC ION | | Authors: | Hakanpaa, J.M, Rouvinen, J. | | Deposit date: | 2006-05-03 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two crystal structures of Trichoderma reesei hydrophobin HFBI--The structure of a protein amphiphile with and without detergent interaction.
Protein Sci., 15, 2006
|
|
