6MJZ
 
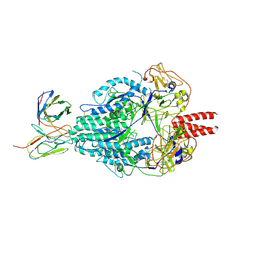 | | Cryo-EM structure of Human Parainfluenza Virus Type 3 (hPIV3) in complex with antibody PIA174 | | Descriptor: | Fusion glycoprotein F0, PIA174 Fab Heavy chain, PIA174 Fab Light chain | | Authors: | Acharya, P, Stewart-Jones, G, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-09-24 | | Release date: | 2018-11-14 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure-based design of a quadrivalent fusion glycoprotein vaccine for human parainfluenza virus types 1-4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8CJZ
 
 | | Carin1 bacteriophage mature capsid | | Descriptor: | Capsid Decoration Protein, Major Capsid Protein, Spike Base Protein | | Authors: | d'Acapito, A, Neumann, E, Schoehn, G. | | Deposit date: | 2023-02-14 | | Release date: | 2023-03-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Study of the Cobetia marina Bacteriophage 1 (Carin-1) by Cryo-EM.
J.Virol., 97, 2023
|
|
8CK0
 
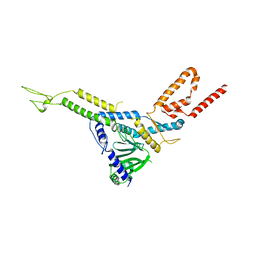 | |
6MWK
 
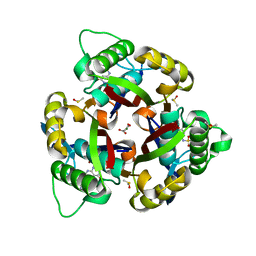 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (IspF) Burkholderia pseudomallei in complex with ligand HGN-0883 | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 4-amino-N-(6-methoxypyrimidin-4-yl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of Burkholderia pseudomallei IspF in complex with sulfapyridine, sulfamonomethoxine, ethoxzolamide and acetazolamide
Acta Crystallogr.,Sect.F, 81, 2025
|
|
6VB1
 
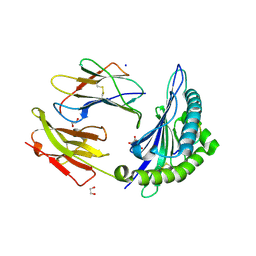 | | HLA-B*15:02 complexed with a synthetic peptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Schutte, R.J, Li, D, Andring, J, McKenna, R, Ostrov, D.A. | | Deposit date: | 2019-12-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | HLA-B*15:02 complexed with a synthetic peptide
To Be Published
|
|
6VB7
 
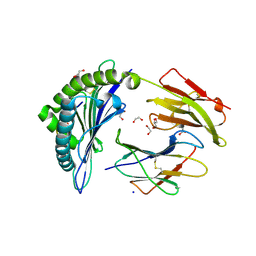 | |
1ETH
 
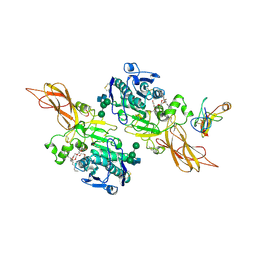 | | TRIACYLGLYCEROL LIPASE/COLIPASE COMPLEX | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Hermoso, J, Pignol, D, Kerfelec, B, Crenon, I, Chapus, C, Fontecilla-Camps, J.C. | | Deposit date: | 1995-09-13 | | Release date: | 1996-12-07 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lipase activation by nonionic detergents. The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether complex.
J.Biol.Chem., 271, 1996
|
|
7QQ1
 
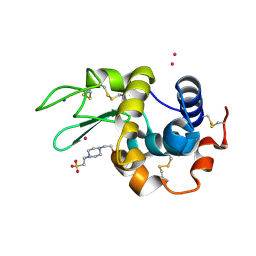 | |
8QT9
 
 | | Cryo-EM structure of stably reduced Streptococcus pneumoniae NADPH oxidase in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FAD-binding FR-type domain-containing protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dubach, V.R.A, San Segundo-Acosta, P, Murphy, B.J. | | Deposit date: | 2023-10-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural and mechanistic insights into Streptococcus pneumoniae NADPH oxidase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QIY
 
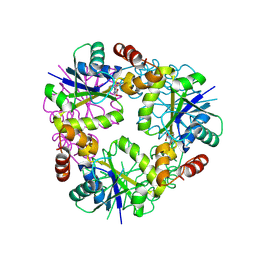 | | Structure of Mycobacterium abscessus Phosphopantetheine adenylyltransferase in complex with inhibitor | | Descriptor: | 1-(2-aminophenyl)-5-(trifluoromethyl)pyrazole-4-carboxylic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, McCarthy, W.J, Coyne, A.G, Blundell, T.L. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.5149 Å) | | Cite: | A Fragment-Based Competitive 19 F LB-NMR Platform For Hotspot-Directed Ligand Profiling.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7R2V
 
 | | Structure of nsp14 from SARS-CoV-2 in complex with SAH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Proofreading exoribonuclease nsp14, ... | | Authors: | Czarna, A, Plewka, J, Kresik, L, Matsuda, A, Abdulkarim, K, Robinson, C, OByrne, S, Cunningham, F, Georgiou, I, Pachota, M, Popowicz, G.M, Wyatt, P.G, Dubin, G, Pyrc, K. | | Deposit date: | 2022-02-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity.
Structure, 30, 2022
|
|
8E8Z
 
 | | 9H2 Fab-Sabin poliovirus 1 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
8VXE
 
 | | Structure of p38 alpha (Mitogen-activated protein kinase 14) complexed with inhibitor 6 | | Descriptor: | (4M)-4-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-1H-pyrrolo[2,3-b]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Blaesse, M, Steinbacher, S, Shaffer, P.L, Sharma, S, Thompson, A.A. | | Deposit date: | 2024-02-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Optimization of Selective and Brain Penetrant CK1 delta Inhibitors for the Treatment of Circadian Disruptions.
Acs Med.Chem.Lett., 15, 2024
|
|
1ELV
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COMPLEMENT C1S PROTEASE | | Descriptor: | 2-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C1S COMPONENT, ... | | Authors: | Gaboriaud, C, Rossi, V, Bally, I, Arlaud, G, Fontecilla-Camps, J.-C. | | Deposit date: | 2000-03-14 | | Release date: | 2001-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the catalytic domain of human complement c1s: a serine protease with a handle.
EMBO J., 19, 2000
|
|
7O7A
 
 | |
6FUN
 
 | |
7A3W
 
 | | Structure of Imine Reductase from Pseudomonas sp. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NAD(P)-dependent oxidoreductase, ... | | Authors: | Cuetos, A, Thorpe, T, Turner, N.J, Grogan, G. | | Deposit date: | 2020-08-18 | | Release date: | 2021-08-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Multifunctional biocatalyst for conjugate reduction and reductive amination.
Nature, 604, 2022
|
|
7QCT
 
 | | PDZ2 of LNX2 with SARS-CoV-2_E PBM complex | | Descriptor: | Envelope small membrane protein, Ligand of Numb protein X 2 | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
8VJ6
 
 | | GluA2 bound to GYKI-52466 and Glutamate, Inhibited State 1 | | Descriptor: | 4-[(5S,8R)-8-methyl-6,7,8,9-tetrahydro-2H,5H-[1,3]dioxolo[4,5-h][2,3]benzodiazepin-5-yl]aniline, Isoform Flip of Glutamate receptor 2 | | Authors: | Hale, W.D, Montano Romero, A, Huganir, R.L, Twomey, E.C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-06-05 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Allosteric competition and inhibition in AMPA receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6MO0
 
 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | Descriptor: | 1-(4-{3-[4-(furan-3-yl)phenyl]-5-[(piperidin-4-yl)methoxy]pyrazin-2-yl}phenyl)methanamine, FLAVIVIRUS_NS2B/Peptidase S7 | | Authors: | Lin, Y.-L, Nie, S, Hua, Y, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | Deposit date: | 2018-10-03 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J. Am. Chem. Soc., 141, 2019
|
|
8T8Z
 
 | | Crystal structure of Terrestrivirus Inositol pyrophosphatase kinase in complex with ADP and myo-(1OH)IP5 | | Descriptor: | (1S,2R,3S,4S,5S,6S)-6-hydroxycyclohexane-1,2,3,4,5-pentayl pentakis[dihydrogen (phosphate)], ADENOSINE-5'-DIPHOSPHATE, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
5OD4
 
 | |
8W0S
 
 | | Human EBP complexed with compound 3a | | Descriptor: | 1-methyl-8-[(oxan-4-yl)methyl]-3-[4-(trifluoromethyl)phenyl]-1,3,8-triazaspiro[4.5]decane-2,4-dione, 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | | Authors: | Sun, D, Masureel, M. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Discovery and Optimization of Selective Brain-Penetrant EBP Inhibitors that Enhance Oligodendrocyte Formation.
J.Med.Chem., 67, 2024
|
|
7QWK
 
 | | GCN2 (EIF2ALPHA KINASE 4, E2AK4) IN COMPLEX WITH COMPOUND 2 | | Descriptor: | (2~{S})-~{N}-[(1~{S})-1-[4-[(6-pyridin-4-ylquinazolin-2-yl)amino]phenyl]ethyl]piperidine-2-carboxamide, DIMETHYL SULFOXIDE, eIF-2-alpha kinase GCN2 | | Authors: | Maia de Oliveira, T. | | Deposit date: | 2022-01-25 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of human GCN2 reveals a parallel, back-to-back kinase dimer with a plastic DFG activation loop motif.
Biochem.J., 477, 2020
|
|
1EKI
 
 | | AVERAGE SOLUTION STRUCTURE OF D(TTGGCCAA)2 BOUND TO CHROMOMYCIN-A3 AND COBALT | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-4-O-acetyl-2,6-dideoxy-beta-D-galactopyranose, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-beta-D-Olivopyranose-(1-3)-beta-D-Olivopyranose, ... | | Authors: | Gochin, M. | | Deposit date: | 2000-03-08 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A high-resolution structure of a DNA-chromomycin-Co(II) complex determined from pseudocontact shifts in nuclear magnetic resonance.
Structure Fold.Des., 8, 2000
|
|
