2R7M
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with AMP | | Descriptor: | 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
3QSL
 
 | | Structure of CAE31940 from Bordetella bronchiseptica RB50 | | Descriptor: | CITRIC ACID, Putative exported protein | | Authors: | Bajor, J, Kagan, O, Chruszcz, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-21 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of pyrimidine/thiamin biosynthesis precursor-like domain-containing protein CAE31940 from proteobacterium Bordetella bronchiseptica RB50, and evolutionary insight into the NMT1/THI5 family.
J Struct Funct Genomics, 15, 2014
|
|
3QO0
 
 | |
2R82
 
 | | Pyruvate phosphate dikinase (PPDK) triple mutant R219E/E271R/S262D adapts a second conformational state | | Descriptor: | Pyruvate, phosphate dikinase, SULFATE ION | | Authors: | Lim, K, Read, R.J, Chen, C.C, Herzberg, O. | | Deposit date: | 2007-09-10 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Swiveling domain mechanism in pyruvate phosphate dikinase.
Biochemistry, 46, 2007
|
|
3EFG
 
 | | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, Protein slyX homolog | | Authors: | Cuff, M.E, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-08 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913
TO BE PUBLISHED
|
|
3QAP
 
 | | Crystal structure of Natronomonas pharaonis sensory rhodopsin II in the ground state | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, EICOSANE, RETINAL, ... | | Authors: | Gushchin, I, Reshetnyak, A, Borshchevskiy, V, Ishchenko, A, Round, E, Grudinin, S, Engelhard, M, Buldt, G, Gordeliy, V. | | Deposit date: | 2011-01-11 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active State of Sensory Rhodopsin II: Structural Determinants for Signal Transfer and Proton Pumping.
J.Mol.Biol., 412, 2011
|
|
2RI7
 
 | |
2RIJ
 
 | | Crystal structure of a putative 2,3,4,5-tetrahydropyridine-2-carboxylate n-succinyltransferase (cj1605c, dapd) from campylobacter jejuni at 1.90 A resolution | | Descriptor: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2007-10-11 | | Release date: | 2007-10-23 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Putative 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase (NP_282733.1) from Campylobacter jejuni at 1.90 A resolution
To be published
|
|
3QN8
 
 | | HIV-1 protease (mutant Q7K L33I L63I) in complex with a novel inhibitor | | Descriptor: | (4aS,7aS)-1,4-bis(3-hydroxybenzyl)hexahydro-1H-pyrrolo[3,4-b]pyrazine-2,3-dione, CHLORIDE ION, Protease | | Authors: | Lindemann, I, Heine, A, Klebe, G. | | Deposit date: | 2011-02-08 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.382 Å) | | Cite: | Novel inhibitors for HIV-1 protease
To be Published
|
|
2R68
 
 | |
3QQ7
 
 | |
3E7D
 
 | |
2R7L
 
 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with ATP and AICAR | | Descriptor: | 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE-5'-TRIPHOSPHATE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
3QJC
 
 | |
3ECJ
 
 | |
3QS3
 
 | |
3QJB
 
 | |
3EI4
 
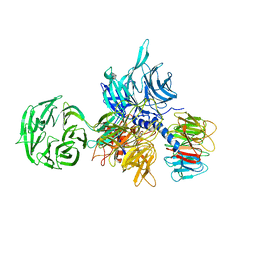 | | Structure of the hsDDB1-hsDDB2 complex | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2 | | Authors: | Scrima, A, Pavletich, N.P, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
3EK6
 
 | |
3EN3
 
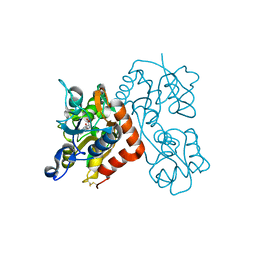 | | Crystal Structure of the GluR4 Ligand-Binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 4,Glutamate receptor | | Authors: | Gill, A, Madden, D.R. | | Deposit date: | 2008-09-25 | | Release date: | 2009-05-19 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Correlating AMPA receptor activation and cleft closure across subunits: crystal structures of the GluR4 ligand-binding domain in complex with full and partial agonists
Biochemistry, 47, 2008
|
|
2R7N
 
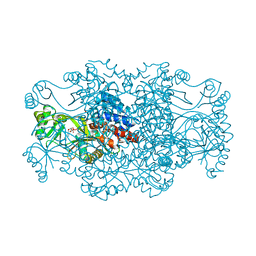 | | Crystal structure of FAICAR synthetase (PurP) from M. jannaschii complexed with ADP and FAICAR | | Descriptor: | 5-(formylamino)-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxamide, 5-formaminoimidazole-4-carboxamide-1-(beta)-D-ribofuranosyl 5'-monophosphate synthetase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
3QNZ
 
 | | Orthorhombic form of IgG1 Fab fragment (in complex with antigenic tubulin peptide) sharing same Fv as IgA | | Descriptor: | Fab fragment of IMMUNOGLOBULIN G1 HEAVY CHAIN, Fab fragment of IMMUNOGLOBULIN G1 LIGHT CHAIN, GLYCEROL, ... | | Authors: | Trajtenberg, F, Correa, A, Buschiazzo, A. | | Deposit date: | 2011-02-09 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a human IgA1 Fab fragment at 1.55 angstrom resolution: potential effect of the constant domains on antigen-affinity modulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3QPJ
 
 | |
2RCJ
 
 | | Solution structure of human Immunoglobulin M | | Descriptor: | IgA1 heavy chain, IgA1 light chain, J chain | | Authors: | Perkins, S.J, Nealis, A.S, Sutton, B.J, Feinstein, A. | | Deposit date: | 2007-09-20 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Solution structure of human and mouse immunoglobulin M by synchrotron X-ray scattering and molecular graphics modelling. A possible mechanism for complement activation.
J.Mol.Biol., 221, 1991
|
|
3ETP
 
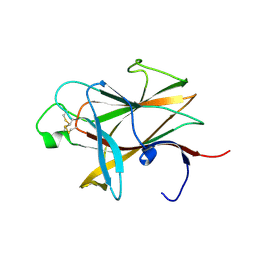 | | The crystal structure of the ligand-binding domain of the EphB2 receptor at 2.0 A resolution | | Descriptor: | Ephrin type-B receptor 2 | | Authors: | Goldgur, Y, Paavilainen, S, Nikolov, D.B, Himanen, J.P. | | Deposit date: | 2008-10-08 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the ligand-binding domain of the EphB2 receptor at 2 A resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
