1RTF
 
 | |
1RX2
 
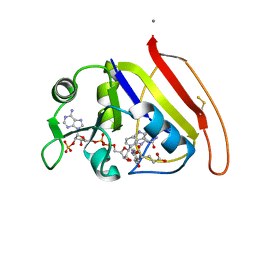 | |
1KRN
 
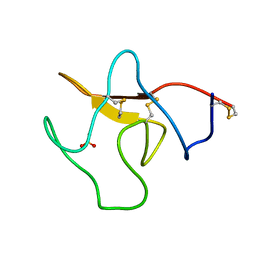 | | STRUCTURE OF KRINGLE 4 AT 4C TEMPERATURE AND 1.67 ANGSTROMS RESOLUTION | | Descriptor: | PLASMINOGEN, SULFATE ION | | Authors: | Stec, B, Teeter, M.M, Whitlow, M, Yamano, A. | | Deposit date: | 1995-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of human plasminogen kringle 4 at 1.68 a and 277 K. A possible structural role of disordered residues.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1IES
 
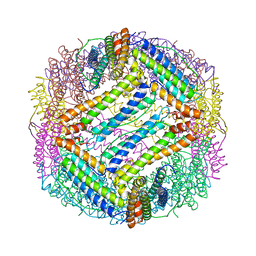 | | TETRAGONAL CRYSTAL STRUCTURE OF NATIVE HORSE SPLEEN FERRITIN | | Descriptor: | CADMIUM ION, FERRITIN | | Authors: | Granier, T, Gallois, B, Dautant, A, Langlois D'Estaintot, B, Precigoux, G. | | Deposit date: | 1996-05-28 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparison of the structures of the cubic and tetragonal forms of horse-spleen apoferritin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1OBR
 
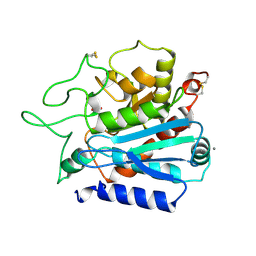 | | CARBOXYPEPTIDASE T | | Descriptor: | CALCIUM ION, CARBOXYPEPTIDASE T, SULFATE ION, ... | | Authors: | Teplyakov, A, Polyakov, K, Obmolova, G, Osterman, A. | | Deposit date: | 1996-06-22 | | Release date: | 1997-01-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of carboxypeptidase T from Thermoactinomyces vulgaris.
Eur.J.Biochem., 208, 1992
|
|
1GBV
 
 | | (ALPHA-OXY, BETA-(C112G)DEOXY) T-STATE HUMAN HEMOGLOBIN | | Descriptor: | HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vasquez, G.B, Ji, X, Pechik, I, Fronticelli, C, Gilliland, G.L. | | Deposit date: | 1995-12-20 | | Release date: | 1997-01-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cysteines beta93 and beta112 as probes of conformational and functional events at the human hemoglobin subunit interfaces.
Biophys.J., 76, 1999
|
|
2HVM
 
 | |
1KCT
 
 | | ALPHA1-ANTITRYPSIN | | Descriptor: | ALPHA1-ANTITRYPSIN | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Crystal structure of an uncleaved alpha 1-antitrypsin reveals the conformation of its inhibitory reactive loop.
FEBS Lett., 377, 1995
|
|
1TVX
 
 | |
1LIT
 
 | | HUMAN LITHOSTATHINE | | Descriptor: | LITHOSTATHINE | | Authors: | Bertrand, J.A, Pignol, D, Bernard, J.-P, Verdier, J.-M, Dagorn, J.-C, Fontacilla-Camps, J.C. | | Deposit date: | 1996-01-17 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of human lithostathine, the pancreatic inhibitor of stone formation.
EMBO J., 15, 1996
|
|
1XBL
 
 | | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | | Descriptor: | DNAJ | | Authors: | Pellecchia, M, Szyperski, T, Wall, D, Georgopoulos, C, Wuthrich, K. | | Deposit date: | 1996-10-07 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone.
J.Mol.Biol., 260, 1996
|
|
1XSM
 
 | | PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE FROM MOUSE | | Descriptor: | FE (III) ION, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Kauppi, B, Nielsen, B.N, Ramaswamy, S, Kjoller-Larsen, I, Thelander, M, Thelander, L, Eklund, H. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of mammalian ribonucleotide reductase protein R2 reveals a more-accessible iron-radical site than Escherichia coli R2.
J.Mol.Biol., 262, 1996
|
|
1QBA
 
 | | BACTERIAL CHITOBIASE, GLYCOSYL HYDROLASE FAMILY 20 | | Descriptor: | CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1FLE
 
 | | CRYSTAL STRUCTURE OF ELAFIN COMPLEXED WITH PORCINE PANCREATIC ELASTASE | | Descriptor: | ELAFIN, ELASTASE | | Authors: | Tsunemi, M, Matsuura, Y, Sakakibara, S, Katsube, Y. | | Deposit date: | 1996-07-04 | | Release date: | 1997-01-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an elastase-specific inhibitor elafin complexed with porcine pancreatic elastase determined at 1.9 A resolution.
Biochemistry, 35, 1996
|
|
1GBU
 
 | | DEOXY (BETA-(C93A,C112G)) HUMAN HEMOGLOBIN | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Vasquez, G.B, Ji, X, Fronticelli, C, Gilliland, G.L. | | Deposit date: | 1996-01-04 | | Release date: | 1997-01-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cysteines beta93 and beta112 as probes of conformational and functional events at the human hemoglobin subunit interfaces.
Biophys.J., 76, 1999
|
|
1GHJ
 
 | | SOLUTION STRUCTURE OF THE LIPOYL DOMAIN OF THE 2-OXOGLUTARATE DEHYDROGENASE COMPLEX FROM AZOTOBACTER VINELAND II, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | E2, THE DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE COMPONENT OF 2-OXOGLUTARATE DEHYDROGENASE COMPLEX | | Authors: | Berg, A, Vervoort, J, De Kok, A. | | Deposit date: | 1996-01-16 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the lipoyl domain of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii.
J.Mol.Biol., 261, 1996
|
|
1IRV
 
 | |
1QPA
 
 | | LIGNIN PEROXIDASE ISOZYME LIP4.65 (PI 4.65) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Choinowski, T.H, Piontek, K. | | Deposit date: | 1996-10-08 | | Release date: | 1997-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of lignin peroxidase at 1.70 A resolution reveals a hydroxy group on the cbeta of tryptophan 171: a novel radical site formed during the redox cycle.
J.Mol.Biol., 286, 1999
|
|
1SME
 
 | | PLASMEPSIN II, A HEMOGLOBIN-DEGRADING ENZYME FROM PLASMODIUM FALCIPARUM, IN COMPLEX WITH PEPSTATIN A | | Descriptor: | PLASMEPSIN II, Pepstatin | | Authors: | Silva, A.M, Lee, A.Y, Gulnik, S.V, Goldberg, D.E, Erickson, J.W. | | Deposit date: | 1996-06-11 | | Release date: | 1997-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and inhibition of plasmepsin II, a hemoglobin-degrading enzyme from Plasmodium falciparum.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1IER
 
 | | CUBIC CRYSTAL STRUCTURE OF NATIVE HORSE SPLEEN FERRITIN | | Descriptor: | CADMIUM ION, FERRITIN | | Authors: | Granier, T, Gallois, B, Dautant, A, Langlois D'Estaintot, B, Precigoux, G. | | Deposit date: | 1996-05-28 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Comparison of the structures of the cubic and tetragonal forms of horse-spleen apoferritin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1GHK
 
 | | SOLUTION STRUCTURE OF THE LIPOYL DOMAIN OF THE 2-OXOGLUTARATE DEHYDROGENASE COMPLEX FROM AZOTOBACTER VINELAND II, NMR, 25 STRUCTURES | | Descriptor: | E2, THE DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE COMPONENT OF 2-OXOGLUTARATE DEHYDROGENASE COMPLEX | | Authors: | Berg, A, Vervoort, J, De Kok, A. | | Deposit date: | 1996-01-16 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the lipoyl domain of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii.
J.Mol.Biol., 261, 1996
|
|
1GTM
 
 | | STRUCTURE OF GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE, SULFATE ION | | Authors: | Yip, K.S.P, Stillman, T.J, Britton, K.L, Pasquo, A, Rice, D.W. | | Deposit date: | 1996-08-22 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures.
Structure, 3, 1995
|
|
1JST
 
 | | PHOSPHORYLATED CYCLIN-DEPENDENT KINASE-2 BOUND TO CYCLIN A | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN A, CYCLIN-DEPENDENT KINASE-2, ... | | Authors: | Russo, A.A, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of cyclin-dependent kinase activation by phosphorylation.
Nat.Struct.Biol., 3, 1996
|
|
1LSQ
 
 | | RIBONUCLEASE A WITH ASN 67 REPLACED BY A BETA-ASPARTYL RESIDUE | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Esposito, L, Sica, F, Vitagliano, L, Zagari, A, Mazzarella, L. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Deamidation in proteins: the crystal structure of bovine pancreatic ribonuclease with an isoaspartyl residue at position 67.
J.Mol.Biol., 257, 1996
|
|
1LCL
 
 | | CHARCOT-LEYDEN CRYSTAL PROTEIN | | Descriptor: | LYSOPHOSPHOLIPASE | | Authors: | Acharya, K.R, Leonidas, D.D. | | Deposit date: | 1996-01-18 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Charcot-Leyden crystal protein, an eosinophil lysophospholipase, identifies it as a new member of the carbohydrate-binding family of galectins.
Structure, 3, 1995
|
|
