2OT4
 
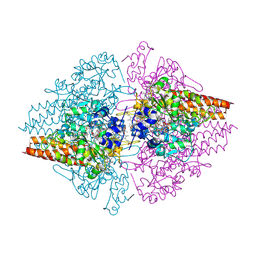 | | Structure of a hexameric multiheme c nitrite reductase from the extremophile bacterium Thiolkalivibrio nitratireducens | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2007-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens.
J.Mol.Biol., 389, 2009
|
|
2ZO5
 
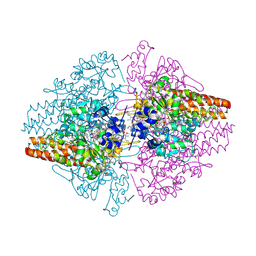 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with azide | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-05 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
1GOY
 
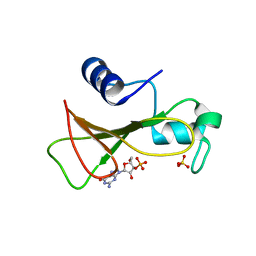 | | HYDROLASE(ENDORIBONUCLEASE)RIBONUCLEASE BI(G SPECIFIC ENDONUCLEASE) (E.C.3.1.27.-) COMPLEXED WITH GUANOSINE-3'-PHOSPHATE (3'-GMP) | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE, SULFATE ION | | Authors: | Polyakov, K.M, Lebedev, A.A, Pavlovsky, A.G, Sanishvili, R.G, Dodson, G.G. | | Deposit date: | 2001-10-26 | | Release date: | 2001-11-29 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Substrate-Free Microbial Ribonuclease Binase and of its Complexes with 3'Gmp and Sulfate Ions
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GOV
 
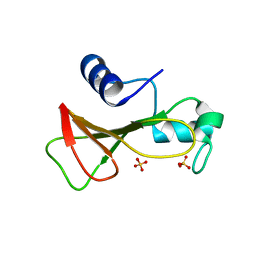 | | RIBONUCLEASE BI(G SPECIFIC ENDONUCLEASE) COMPLEXED WITH SULFATE IONS | | Descriptor: | RIBONUCLEASE, SULFATE ION | | Authors: | Polyakov, K.M, Lebedev, A.A, Pavlovsky, A.G, Sanishvili, R.G, Dodson, G.G. | | Deposit date: | 2001-10-26 | | Release date: | 2001-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Substrate-Free Microbial Ribonuclease Binase and of its Complexes with 3'Gmp and Sulfate Ions
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5OEX
 
 | | Complex with iodine ion for thiocyanate dehydrogenase from Thioalkalivibrio paradoxus | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Tsallagov, S.I, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2017-07-10 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of a novel copper containing enzyme - THIOCYANATE DEHYDROGENASE.
To Be Published
|
|
7Q2W
 
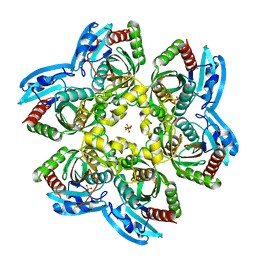 | |
3D1I
 
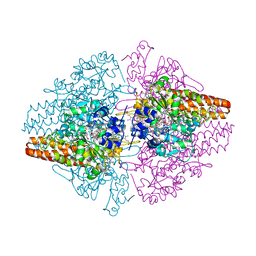 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in a complex with nitrite | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens
J.Mol.Biol., 389, 2009
|
|
7Q30
 
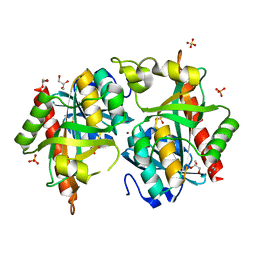 | |
7Q1I
 
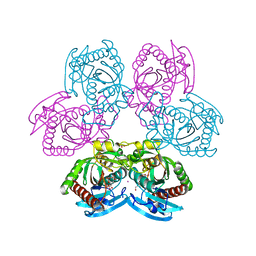 | |
8RXU
 
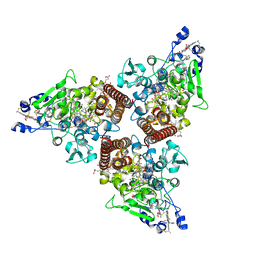 | | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in space group P21 | | Descriptor: | CALCIUM ION, HEME C, Octaheme nitrite reductase, ... | | Authors: | Polyakov, K.M, Safonova, T.N, Osipov, E, Popov, A.N, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-02-08 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in space group P21
To Be Published
|
|
3TTB
 
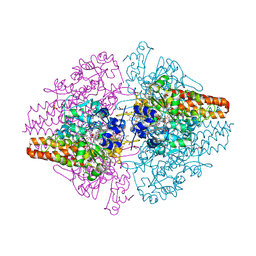 | | Structure of the Thioalkalivibrio paradoxus cytochrome c nitrite reductase in complex with sulfite | | Descriptor: | CALCIUM ION, COBALT (II) ION, Eight-heme nitrite reductase, ... | | Authors: | Polyakov, K.M, Trofimov, A.A, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | Deposit date: | 2011-09-14 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative structural and functional analysis of two octaheme nitrite reductases from closely related Thioalkalivibrio species.
Febs J., 279, 2012
|
|
8RV0
 
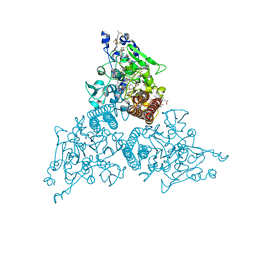 | | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in complex with nitrite | | Descriptor: | CALCIUM ION, HEME C, NITRIC OXIDE, ... | | Authors: | Polyakov, K.M, Safonova, T.N, Osipov, E, Popov, A.N, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-01-31 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in complex with nitrite
To Be Published
|
|
8RVM
 
 | | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in space group P63 | | Descriptor: | CALCIUM ION, HEME C, Octaheme nitrite c cytochrome c reductase, ... | | Authors: | Polyakov, K.M, Safoonova, T.N, Osipov, E, Popov, A.N, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-02-01 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of octaheme nitrite reductase from Trichlorobacter ammonificans in space group P63
To Be Published
|
|
1GOU
 
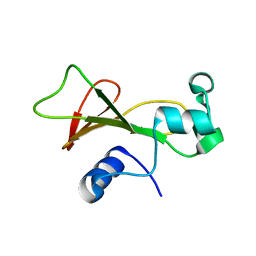 | |
6I3Q
 
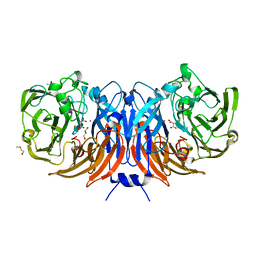 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus complex with acetate ions. | | Descriptor: | ACETATE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Polyakov, K.M, Popov, A.N, Tikhkonova, T.V, Popov, V.O, Trofimov, A.A. | | Deposit date: | 2018-11-07 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6G5M
 
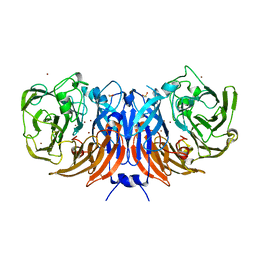 | |
3FPX
 
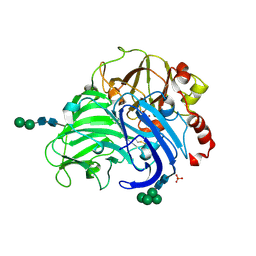 | | Native fungus laccase from Trametes hirsuta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Fedorova, T.V, Stepanova, E.V, Cherkashin, E.A, Kurzeev, S.A, Strokopytov, B.V, Lamzin, V.S, Koroleva, O.V. | | Deposit date: | 2009-01-06 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of native laccase from Coriolus hirsutus at 1.8 A resolution
To be Published
|
|
3SXQ
 
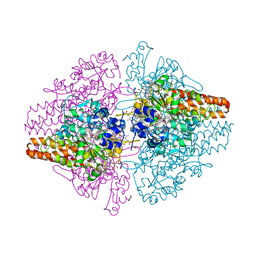 | | Structure of a hexameric multiheme c nitrite reductase from the extremophile bacterium Thiolkalivibrio paradoxus | | Descriptor: | CALCIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Polyakov, K.M, Trofimov, A.A, Tikhonova, T.V, Tikhonov, A.V, Boyko, K.M, Popov, V.O. | | Deposit date: | 2011-07-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative structural and functional analysis of two octaheme nitrite reductases from closely related Thioalkalivibrio species.
Febs J., 279, 2012
|
|
4JHV
 
 | | T2-depleted laccase from Coriolopsis caperata | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Fedorova, T.V, Glazunova, O.A, Kurzeev, S.A, Maloshenok, L.G, Lamzin, V.S, Koroleva, O.V. | | Deposit date: | 2013-03-05 | | Release date: | 2014-04-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elucidation of the crystal structure of Coriolopsis caperata laccase: restoration of the structure and activity of the native enzyme from the T2-depleted form by copper ions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5E9N
 
 | | Steccherinum murashkinskyi laccase at 0.95 resolution | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Glazunova, O.A, Fedorova, T.V, Koroleva, O.V. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structure-function study of two new middle-redox potential laccases from basidiomycetes Antrodiella faginea and Steccherinum murashkinskyi.
Int. J. Biol. Macromol., 118, 2018
|
|
5EHF
 
 | | Laccase from Antrodiella faginea | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Glazunova, O.A, Fedorova, T.V, Dorovatovskii, P.V, Koroleva, O.V. | | Deposit date: | 2015-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-function study of two new middle-redox potential laccases from basidiomycetes Antrodiella faginea and Steccherinum murashkinskyi.
Int. J. Biol. Macromol., 118, 2018
|
|
3V9C
 
 | | Type-2 Cu-depleted fungus laccase from Trametes hirsuta at low dose of ionization radiation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Fedorova, T.V, Kurzeev, S.A, Popov, A.N, Lamzin, V.S, Koroleva, O.V. | | Deposit date: | 2011-12-26 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of T2-depleted fungal laccase from Trametes hirsuta 072 at low and high dose of ionization radiation
To be Published
|
|
4HAA
 
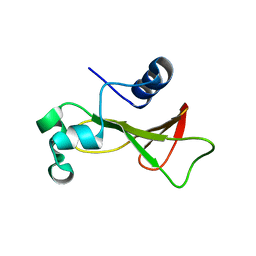 | | Structure of Ribonuclease Binase Glu43Ala/Phe81Ala Mutant | | Descriptor: | Ribonuclease | | Authors: | Polyakov, K.M, Trofimov, A.A, Mitchevich, V.A, Dorovatovskii, P.V, Schulga, A.A, Makarov, A.A, Tkach, E.N, Goncharuk, D.A. | | Deposit date: | 2012-09-26 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and functional studies of the ribonuclease binase Glu43Ala/Phe81Ala mutant.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JHU
 
 | | T2-depleted laccase from Coriolopsis caperata soaked with CuCl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, LACCASE, ... | | Authors: | Polyakov, K.M, Fedorova, T.V, Glazunova, O.A, Kurzeev, S.A, Maloshenok, L.G, Koroleva, O.A. | | Deposit date: | 2013-03-05 | | Release date: | 2014-04-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Elucidation of the crystal structure of Coriolopsis caperata laccase: restoration of the structure and activity of the native enzyme from the T2-depleted form by copper ions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3PXL
 
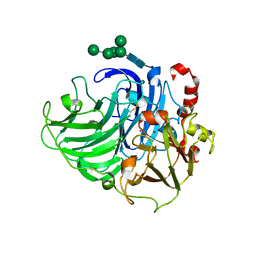 | | Type-2 Cu-depleted fungus laccase from Trametes hirsuta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Fedorova, T.V, Kurzeev, S.A, Popov, A.N, Lamzin, V.S, Koroleva, O.V. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of T2-depleted fungal laccase from Trametes hirsuta at low and high dose of ionization radiation.
To be Published
|
|
