1EAU
 
 | | NONPEPTIDIC INHIBITORS OF HUMAN LEUKOCYTE ELASTASE. 6. DESIGN OF A POTENT, INTRATRACHEALLY ACTIVE, PYRIDONE-BASED TRIFLUOROMETHYL KETONE | | Descriptor: | 2-[5-AMINO-6-OXO-2-(2-THIENYL)-1,6-DIHYDROPYRIMIDIN-1-YL)-N-[3,3-DIFLUORO -1-ISOPROPYL-2-OXO-3-(N-(2-MORPHOLINO ETHYL)CARBAMOYL]PROPYL]ACETAMIDE, PORCINE PANCREATIC ELASTASE, SODIUM ION, ... | | Authors: | Ceccarelli, C. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nonpeptidic inhibitors of human leukocyte elastase. 6. Design of a potent, intratracheally active, pyridone-based trifluoromethyl ketone.
J.Med.Chem., 38, 1995
|
|
8EN4
 
 | | Structure of GII.4 norovirus in complex with Nanobody 53 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody 53, VP1 | | Authors: | Kher, G, Sabin, C, Koromyslova, A, Pancera, M, Hansman, G. | | Deposit date: | 2022-09-28 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Direct Blockade of the Norovirus Histo-Blood Group Antigen Binding Pocket by Nanobodies.
J.Virol., 97, 2023
|
|
1EAT
 
 | | NONPEPTIDIC INHIBITORS OF HUMAN LEUKOCYTE ELASTASE. 5. DESIGN, SYNTHESIS, AND X-RAY CRYSTALLOGRAPHY OF A SERIES OF ORALLY ACTIVE 5-AMINO-PYRIMIDIN-6-ONE-CONTAINING TRIFLUOROMETHYLKETONES | | Descriptor: | 2-[5-METHANESULFONYLAMINO-2-(4-AMINOPHENYL)-6-OXO-1,6-DIHYDRO-1-PYRIMIDINYL]-N-(3,3,3-TRIFLUORO-1-ISOPROPYL-2-OXOPROPYL)ACETAMIDE, PORCINE PANCREATIC ELASTASE, SODIUM ION, ... | | Authors: | Ceccarelli, C. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nonpeptidic inhibitors of human leukocyte elastase. 5. Design, synthesis, and X-ray crystallography of a series of orally active 5-aminopyrimidin-6-one-containing trifluoromethyl ketones.
J.Med.Chem., 38, 1995
|
|
7O9T
 
 | |
8EN5
 
 | | Structure of GII.4 norovirus in complex with Nanobody 56 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GII.4 P domain, ... | | Authors: | Kher, G, Sabin, C, Pancera, M, Koromyslova, A, Hansman, G. | | Deposit date: | 2022-09-28 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct Blockade of the Norovirus Histo-Blood Group Antigen Binding Pocket by Nanobodies.
J.Virol., 97, 2023
|
|
7OAS
 
 | | Structural basis for targeted p97 remodeling by ASPL as prerequisite for p97 trimethylation by METTL21D | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | | Authors: | Petrovic, S, Heinemann, U, Roske, Y. | | Deposit date: | 2021-04-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Structural remodeling of AAA+ ATPase p97 by adaptor protein ASPL facilitates posttranslational methylation by METTL21D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8Z9Z
 
 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-24 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
8EMY
 
 | | Structure of GII.4 norovirus in complex with Nanobody 82 | | Descriptor: | 1,2-ETHANEDIOL, GII.4 P domain, Nanobody 82 | | Authors: | Kher, G, Sabin, C, Pancera, M, Koromyslova, A, Hansman, G. | | Deposit date: | 2022-09-28 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Direct Blockade of the Norovirus Histo-Blood Group Antigen Binding Pocket by Nanobodies.
J.Virol., 97, 2023
|
|
8ZAV
 
 | | alcohol dehydrogenases KpADH mutant - S9Y/F161K | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Ni, Y, Xu, G.C. | | Deposit date: | 2024-04-25 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering alcohol dehydrogenases KpADH for enhanced organic-solvent tolerance and its molecular mechanisms
To Be Published
|
|
1F40
 
 | | SOLUTION STRUCTURE OF FKBP12 COMPLEXED WITH GPI-1046, A NEUROTROPHIC LIGAND | | Descriptor: | (2S)-[3-PYRIDYL-1-PROPYL]-1-[3,3-DIMETHYL-1,2-DIOXOPENTYL]-2-PYRROLIDINECARBOXYLATE, FK506 BINDING PROTEIN (FKBP12) | | Authors: | Sich, C, Improta, S, Cowley, D.J, Guenet, C, Merly, J.P, Teufel, M, Saudek, V. | | Deposit date: | 2000-06-07 | | Release date: | 2000-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a neurotrophic ligand bound to FKBP12 and its effects on protein dynamics.
Eur.J.Biochem., 267, 2000
|
|
7P36
 
 | | X-ray structure of Lactobacillus kefir alcohol dehydrogenase (wild type) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bischoff, D, Walla, B, Janowski, R, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Transfer of a Rational Crystal Contact Engineering Strategy between Diverse Alcohol Dehydrogenases
Crystals, 11, 2021
|
|
8CLV
 
 | |
8F7O
 
 | | BRAF kinase in complex with TAK580 (tovorafenib) | | Descriptor: | 6-amino-5-chloro-N-[(1R)-1-(5-{[5-chloro-4-(trifluoromethyl)pyridin-2-yl]carbamoyl}-1,3-thiazol-2-yl)ethyl]pyrimidine-4-carboxamide, Serine/threonine-protein kinase B-raf | | Authors: | Tkacik, E, Li, K, Gonzalez Del-Pino, G, Eck, M.J. | | Deposit date: | 2022-11-20 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Structure and RAF family kinase isoform selectivity of type II RAF inhibitors tovorafenib and naporafenib.
J.Biol.Chem., 299, 2023
|
|
7P7Y
 
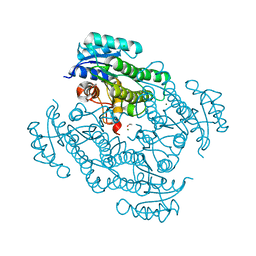 | | X-ray structure of Lactobacillus kefir alcohol dehydrogenase mutant Q126K | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bischoff, D, Walla, B, Janowski, R, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Transfer of a Rational Crystal Contact Engineering Strategy between Diverse Alcohol Dehydrogenases
Crystals, 11, 2021
|
|
8CJW
 
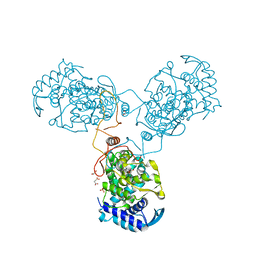 | | Nucleoprotein Thogotovirus delta188-196 | | Descriptor: | 1,2-ETHANEDIOL, Nucleoprotein, PHOSPHATE ION, ... | | Authors: | Dick, A, Roske, Y. | | Deposit date: | 2023-02-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural characterization of Thogoto Virus nucleoprotein provides insights into viral RNA encapsidation and RNP assembly.
Structure, 32, 2024
|
|
8AJ7
 
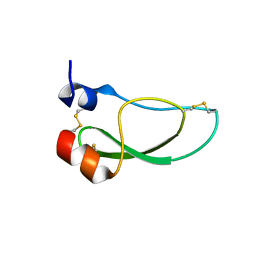 | | Kunitz domain of Amblyomin-X | | Descriptor: | 1,2-ETHANEDIOL, Kunitz domain of Amblyomin-X | | Authors: | Ciccone, L, Servent, D, Stura, E.A. | | Deposit date: | 2022-07-27 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional properties of the Kunitz-type and C-terminal domains of Amblyomin-X supporting its antitumor activity.
Front Mol Biosci, 10, 2023
|
|
5WJO
 
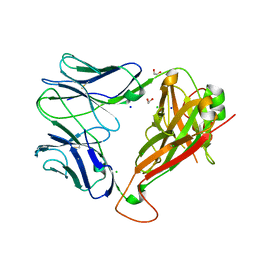 | | Crystal structure of the unliganded PG90 TCR | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PG90 TCR alpha chain, ... | | Authors: | Shahine, A, Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A molecular basis of human T cell receptor autoreactivity toward self-phospholipids.
Sci Immunol, 2, 2017
|
|
8WAF
 
 | | Crystal structure of the C-terminal fragment (residues 756-982 with the C864S mutation) of Arabidopsis thaliana CHUP1 | | Descriptor: | Protein CHUP1, chloroplastic | | Authors: | Shimada, A, Takano, A, Nakamura, Y, Kohda, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CHLOROPLAST UNUSUAL POSITIONING 1 is a plant-specific actin polymerization factor regulating chloroplast movement.
Plant Cell, 36, 2024
|
|
6WAS
 
 | | Structure of D19.PA8 Fab in complex with 1FD6 16055 V1V2 scaffold | | Descriptor: | 1FD6 16055 V1V2 scaffold, 2-acetamido-2-deoxy-beta-D-glucopyranose, GN1_PA8 Fab Heavy chain, ... | | Authors: | Singh, S, Liban, T.J, Pancera, M. | | Deposit date: | 2020-03-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structurally related but genetically unrelated antibody lineages converge on an immunodominant HIV-1 Env neutralizing determinant following trimer immunization.
Plos Pathog., 17, 2021
|
|
8KFV
 
 | | Crystal structure of ZmMOC1 K229A in complex with a nicked Holliday junction soaked in Mn2+ for 180 seconds | | Descriptor: | 1,2-ETHANEDIOL, DNA (25-MER), DNA (33-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8YVJ
 
 | | Crystal structure of the C. difficile toxin A CROPs domain fragment 2592-2710 bound to H5.2 nanobody | | Descriptor: | 1,2-ETHANEDIOL, H5.2 nanobody (VHH), PHOSPHATE ION, ... | | Authors: | Sluchanko, N.N, Varfolomeeva, L.A, Shcheblyakov, D.V, Belyi, Y.F, Logunov, D.Y, Gintsburg, A.L, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-03-28 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insight into recognition of Clostridioides difficile toxin A by novel neutralizing nanobodies targeting QTIN-like motifs within its receptor-binding domain.
Int.J.Biol.Macromol., 283, 2024
|
|
7RNT
 
 | | CRYSTAL STRUCTURE OF THE TYR45TRP MUTANT OF RIBONUCLEASE T1 IN A COMPLEX WITH 2'-ADENYLIC ACID | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, CALCIUM ION, RIBONUCLEASE T1 | | Authors: | Koellner, G, Grunert, H.-P, Landt, O, Saenger, W. | | Deposit date: | 1991-08-20 | | Release date: | 1993-01-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Tyr45Trp mutant of ribonuclease T1 in a complex with 2'-adenylic acid.
Eur.J.Biochem., 201, 1991
|
|
7OYF
 
 | |
7OYH
 
 | |
6WKH
 
 | | Crystal structure of pentalenene synthase mutant F76W complexed with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Pentalenene synthase | | Authors: | Prem Kumar, R, Matos, J.O, Oprian, D.D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism Underlying Anti-Markovnikov Addition in the Reaction of Pentalenene Synthase.
Biochemistry, 59, 2020
|
|
