2CJU
 
 | |
2COQ
 
 | |
1SM3
 
 | | CRYSTAL STRUCTURE OF THE TUMOR SPECIFIC ANTIBODY SM3 COMPLEX WITH ITS PEPTIDE EPITOPE | | Descriptor: | CADMIUM ION, CHLORIDE ION, PEPTIDE EPITOPE, ... | | Authors: | Dokurno, P, Bates, P.A, Band, H.A, Stewart, L.M.D, Lally, J.M, Burchell, J.M, Taylor-Papadimitriou, J, Sternberg, M.J.E, Snary, D, Freemont, P.S. | | Deposit date: | 1997-12-23 | | Release date: | 1999-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure at 1.95 A resolution of the breast tumour-specific antibody SM3 complexed with its peptide epitope reveals novel hypervariable loop recognition.
J.Mol.Biol., 284, 1998
|
|
1TCR
 
 | | MURINE T-CELL ANTIGEN RECEPTOR 2C CLONE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garcia, K.C, Degano, M, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 1996-09-12 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex.
Science, 274, 1996
|
|
1SVZ
 
 | | Crystal structure of the single-chain Fv fragment 1696 in complex with the epitope peptide corresponding to N-terminus of HIV-2 protease | | Descriptor: | epitope peptide corresponding to N-terminus of HIV-2 protease, single-chain Fv fragment 1696 | | Authors: | Rezacova, P, Brynda, J, Lescar, J, Bentley, G.A, Fabry, M, Horejsi, M, Sedlacek, J. | | Deposit date: | 2004-03-30 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of a cross-reaction complex between an anti-HIV-1 protease antibody and an HIV-2 protease peptide
J.Struct.Biol., 149, 2005
|
|
2APW
 
 | | Crystal Structure of the G17E/A52V/S54N/K66E/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APV
 
 | | Crystal Structure of the G17E/A52V/S54N/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2ARJ
 
 | | CD8alpha-alpha in complex with YTS 105.18 Fab | | Descriptor: | T-cell surface glycoprotein CD8 alpha chain, YTS 105.18 antigen binding region Heavy chain, YTS 105.18 antigen binding region Light chain | | Authors: | Shore, D.A, Teyton, L, Dwek, R.A, Rudd, P.M, Wilson, I.A. | | Deposit date: | 2005-08-19 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of the TCR co-receptor CD8alphaalpha in complex with monoclonal antibody YTS 105.18 Fab fragment at 2.88 A resolution.
J.Mol.Biol., 358, 2006
|
|
1PEW
 
 | | High Resolution Crystal Structure of Jto2, a mutant of the non-amyloidogenic Lamba6 Light Chain, Jto | | Descriptor: | CADMIUM ION, Jto2, a LAMBDA-6 TYPE IMMUNOGLOBULIN LIGHT CHAIN, ... | | Authors: | Dealwis, C, Gupta, V, Wilkerson, M. | | Deposit date: | 2003-05-22 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of light chain amyloidogenicity: comparison of the thermodynamic properties, fibrillogenic potential and tertiary structural features of four V(lambda)6 proteins
J.Mol.Recog., 17, 2004
|
|
1T2J
 
 | |
2OR8
 
 | | Tim-1 | | Descriptor: | ACETATE ION, Hepatitis A virus cellular receptor 1 homolog | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Casasnovas, J.M. | | Deposit date: | 2007-02-02 | | Release date: | 2007-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Receptors 1 and 2 Reveal Mechanisms for Regulation of Immune Responses by the TIM Receptor Family.
Immunity, 26, 2007
|
|
5XXY
 
 | |
5YD4
 
 | |
2P1Y
 
 | |
2OR7
 
 | | Tim-2 | | Descriptor: | ACETATE ION, T-cell immunoglobulin and mucin domain-containing protein 2 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Casasnovas, J.M. | | Deposit date: | 2007-02-02 | | Release date: | 2007-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Receptors 1 and 2 Reveal Mechanisms for Regulation of Immune Responses by the TIM Receptor Family.
Immunity, 26, 2007
|
|
2OTU
 
 | | Crystal structure of Fv polyglutamine complex | | Descriptor: | Fv heavy chain variable domain, Fv light chain variable domain, peptide antigen | | Authors: | Li, P. | | Deposit date: | 2007-02-09 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Implications of the structure of a poly-Gln/anti-poly-Gln complex for disease progression and therapy
To be Published
|
|
2OYP
 
 | | T Cell Immunoglobulin Mucin-3 Crystal Structure Revealed a Galectin-9-independent Binding Surface | | Descriptor: | Hepatitis A virus cellular receptor 2, SULFATE ION | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | T cell immunoglobulin mucin-3 crystal structure reveals a galectin-9-independent ligand-binding surface
Immunity, 26, 2007
|
|
5YD5
 
 | |
5YD3
 
 | | Crystal structure of the scFv antibody 4B08 with epitope peptide | | Descriptor: | Epitope peptide, GLYCEROL, SULFATE ION, ... | | Authors: | Caaveiro, J.M.M, Miyanabe, K, Tsumoto, K. | | Deposit date: | 2017-09-11 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Intramolecular H-bonds govern the recognition of a flexible peptide by an antibody
J. Biochem., 164, 2018
|
|
5Z11
 
 | | Crystal Structure of Grass Carp CD8 alpha alpha Homodimers | | Descriptor: | CD8 alpha chain | | Authors: | Wang, J. | | Deposit date: | 2017-12-22 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Grass Carp CD8 alpha
alpha Homodimers Indicates a Dramatic Evolution of CD8 from Ectotherms to Endotherms.
To Be Published
|
|
2Q20
 
 | |
2QSQ
 
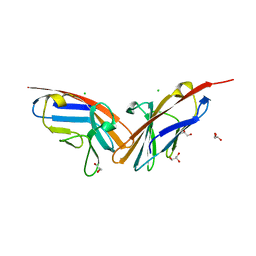 | | Crystal structure of the N-terminal domain of carcinoembryonic antigen (CEA) | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 5, GLYCEROL | | Authors: | Le Trong, I, Korotkova, N, Moseley, S.L, Stenkamp, R.E. | | Deposit date: | 2007-07-31 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding of Dr adhesins of Escherichia coli to carcinoembryonic antigen triggers receptor dissociation.
Mol.Microbiol., 67, 2008
|
|
2OCW
 
 | |
6AW1
 
 | | Crystal structure of CEACAM3 | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
2Q87
 
 | |
