6HQR
 
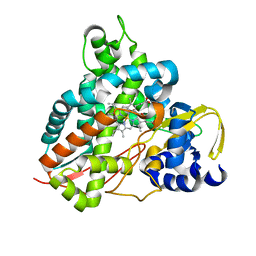 | | Crystal structure of GcoA F169H bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5HCK
 
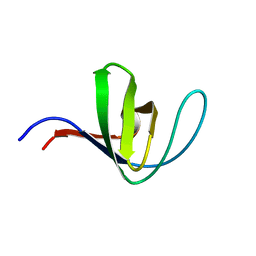 | | HUMAN HCK SH3 DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HEMATOPOIETIC CELL KINASE | | Authors: | Horita, D.A, Baldisseri, D.M, Zhang, W, Altieri, A.S, Smithgall, T.E, Gmeiner, W.H, Byrd, R.A. | | Deposit date: | 1998-03-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human Hck SH3 domain and identification of its ligand binding site.
J.Mol.Biol., 278, 1998
|
|
5L15
 
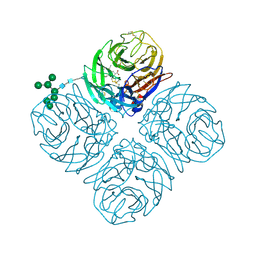 | | The crystal structure of neuraminidase in complex with oseltamivir from A/Shanghai/2/2013 (H7N9) influenza virus | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, H, Stevens, J. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Drug Susceptibility Evaluation of an Influenza A(H7N9) Virus by Analyzing Recombinant Neuraminidase Proteins.
J. Infect. Dis., 216, 2017
|
|
5L1V
 
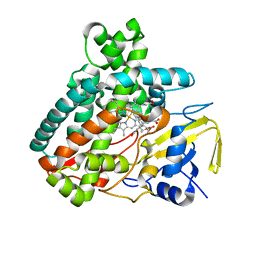 | |
5HBI
 
 | | SCAPHARCA DIMERIC HEMOGLOBIN, MUTANT T72I, CO-LIGANDED FORM | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Royer Junior, W.E. | | Deposit date: | 1998-06-24 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mutational destabilization of the critical interface water cluster in Scapharca dimeric hemoglobin: structural basis for altered allosteric activity.
J.Mol.Biol., 284, 1998
|
|
7BYP
 
 | | Lysozyme structure SASE1 from SASE mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
6HGK
 
 | |
7BYO
 
 | | Lysozyme structure SS1 from SS mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
5I4C
 
 | | Crystal structure of non-phosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli | | Descriptor: | Transcriptional regulatory protein RcsB | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Ruan, J, Pshenychnyi, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of nonphosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli.
Protein Sci., 25, 2016
|
|
6HIP
 
 | | Structure of SPF45 UHM bound to HIV-1 Rev ULM | | Descriptor: | HIV-1 Rev (41-49), SODIUM ION, Splicing factor 45, ... | | Authors: | Pabis, M, Corsini, L, Sattler, M. | | Deposit date: | 2018-08-30 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Modulation of HIV-1 gene expression by binding of a ULM motif in the Rev protein to UHM-containing splicing factors.
Nucleic Acids Res., 47, 2019
|
|
5L92
 
 | |
6HTU
 
 | | Structure of hStau1 dsRBD3-4 in complex with ARF1 RNA | | Descriptor: | Double-stranded RNA-binding protein Staufen homolog 1, RNA (19-MER) | | Authors: | Emmerich, C, Lazzaretti, D, Bandholz-Cajamarca, L, Bono, F. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | The crystal structure of Staufen1 in complex with a physiological RNA sheds light on substrate selectivity.
Life Sci Alliance, 1, 2018
|
|
5KXW
 
 | | Hen Egg White Lysozyme at 278K, Data set 6 | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5I54
 
 | |
5I4W
 
 | | Exploring the onset of lysozyme denaturation by urea | | Descriptor: | Lysozyme C | | Authors: | Hosur, M.V, Raskar, T. | | Deposit date: | 2016-02-13 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-dependent X-ray diffraction studies on urea/hen egg white lysozyme complexes reveal structural changes that indicate onset of denaturation
Sci Rep, 6, 2016
|
|
5L14
 
 | |
6HWV
 
 | | Crystal structure of p38alpha in complex with a photoswitchable 2-Azoimidazol-based Inhibitor (compound 3) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[5-(4-fluorophenyl)-2-[(~{E})-(4-fluorophenyl)diazenyl]-3-methyl-imidazol-4-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Mueller, M.P, Rauh, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
5I5Q
 
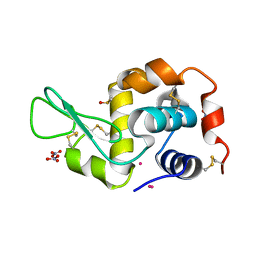 | | Re refinement of 4mwn. | | Descriptor: | DIMETHYL SULFOXIDE, Lysozyme C, NITRATE ION, ... | | Authors: | Helliwell, J.R. | | Deposit date: | 2016-02-15 | | Release date: | 2016-05-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Comment on "Structural dynamics of cisplatin binding to histidine in a protein" [Struct. Dyn. 1, 034701 (2014)].
Struct Dyn, 3, 2016
|
|
7BU4
 
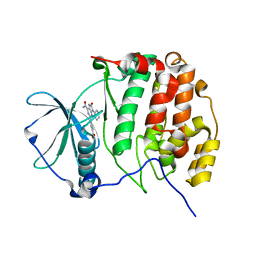 | | Crystal structure of CK2a1 complexed with KY49 | | Descriptor: | 4-(6-aminocarbonyl-8-oxidanylidene-9-phenyl-7H-purin-2-yl)benzoic acid, Casein Kinase 2 subunit alpha | | Authors: | Tsuyuguchi, M, Kinoshita, T. | | Deposit date: | 2020-04-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.70227313 Å) | | Cite: | Design, synthesis and SAR studies of protein kinase CK2 inhibitors with a purine scaffold
To Be Published
|
|
5L1U
 
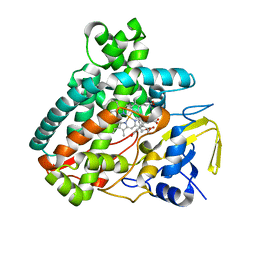 | |
5L23
 
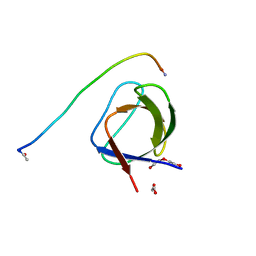 | |
5KXZ
 
 | | Hen Egg White Lysozyme at 278K, Data set 9 | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5L2T
 
 | | The X-ray co-crystal structure of human CDK6 and Ribociclib. | | Descriptor: | 7-cyclopentyl-N,N-dimethyl-2-{[5-(piperazin-1-yl)pyridin-2-yl]amino}-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Cyclin-dependent kinase 6 | | Authors: | Chen, P, Ferre, R.A, Deihl, W, Yu, X, He, Y.-A. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance.
Mol.Cancer Ther., 15, 2016
|
|
6HOR
 
 | | Human protein kinase CK2 alpha in complex with feruloylmethane | | Descriptor: | (~{E})-4-(3-methoxy-4-oxidanyl-phenyl)but-3-en-2-one, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Battistutta, R, Lolli, G. | | Deposit date: | 2018-09-18 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and cellular mechanism of protein kinase CK2 inhibition by deceptive curcumin.
Febs J., 287, 2020
|
|
5I3O
 
 | | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor | | Descriptor: | BMP-2-inducible protein kinase, N-(6-{3-[(dimethylsulfamoyl)amino]phenyl}-1H-indazol-3-yl)cyclopropanecarboxamide, SULFATE ION | | Authors: | Counago, R.M, Sorrell, F.J, Krojer, T, Savitsky, P, Elkins, J.M, Axtman, A, Drewry, D, Wells, C, Zhang, C, Zuercher, W, Willson, T.M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Arruda, P, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor
To Be Published
|
|
