4UUG
 
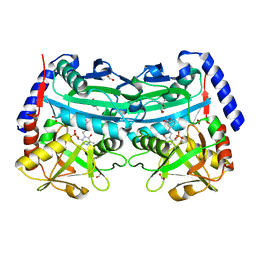 | | The (R)-selective amine transaminase from Aspergillus fumigatus with inhibitor bound | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, ... | | Authors: | Thomsen, M, Hinrichs, W. | | Deposit date: | 2014-07-28 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Characterization of the Dual Substrate Recognition of the (R)-Selective Amine Transaminase from Aspergillus Fumigatus
FEBS J., 282, 2015
|
|
4UWI
 
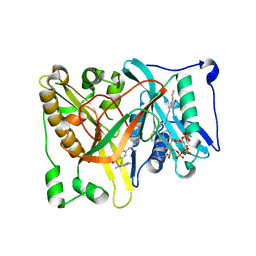 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a pyrazole sulphonamide ligand | | Descriptor: | 2,6-dichloro-4-[3-(4-methylpiperazin-1-yl)propyl]-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Norcross, N.R, Thompson, S, Harrison, J.R, Smith, V.C, Torrie, L.S, McElroy, S.P, Hallyburton, I, Norval, S, Stojanovski, L, Simeons, F.R.C, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lead optimization of a pyrazole sulfonamide series of Trypanosoma brucei N-myristoyltransferase inhibitors: identification and evaluation of CNS penetrant compounds as potential treatments for stage 2 human African trypanosomiasis.
J. Med. Chem., 57, 2014
|
|
4UWJ
 
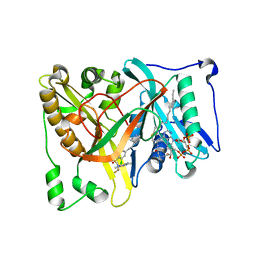 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a capped pyrazole sulphonamide ligand | | Descriptor: | 2,6-dichloro-N-(difluoromethyl)-4-[3-(piperidin-4-yl)propyl]-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Norcross, N.R, Thompson, S, Harrison, J.R, Smith, V.C, Torrie, L.S, McElroy, S.P, Hallyburton, I, Norval, S, Stojanovski, L, Simeons, F.R.C, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lead optimization of a pyrazole sulfonamide series of Trypanosoma brucei N-myristoyltransferase inhibitors: identification and evaluation of CNS penetrant compounds as potential treatments for stage 2 human African trypanosomiasis.
J. Med. Chem., 57, 2014
|
|
7UMH
 
 | | Energetic robustness to large scale structural dynamics in a photosynthetic supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Harris, D, Toporik, H, Schlau-Cohen, G.S, Mazor, Y. | | Deposit date: | 2022-04-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Energetic robustness to large scale structural fluctuations in a photosynthetic supercomplex.
Nat Commun, 14, 2023
|
|
8POF
 
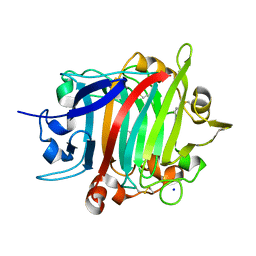 | | The crystal structure of RsSymEG1 reveals a unique form of smaller GH7 endoglucanases alongside GH7 cellobiohydrolases in protist symbionts of termites | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative glycosyl hydrolase family7, SODIUM ION | | Authors: | Haataja, T, Sandgren, M, Hansson, H, Stahlberg, J. | | Deposit date: | 2023-07-04 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of RsSymEG1 reveals a unique form of smaller GH7 endoglucanases alongside GH7 cellobiohydrolases in protist symbionts of termites.
Febs J., 291, 2024
|
|
1HJS
 
 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-1,4-GALACTANASE, ... | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
1HJU
 
 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-1,4-GALACTANASE, ... | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
1HJQ
 
 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-1,4-GALACTANASE | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-07-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
8PMS
 
 | | NADase from Aspergillus fumigatus with replaced C-terminus from Neurospora crassa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Ferrario, E, Stromland, O, Ziegler, M. | | Deposit date: | 2023-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel Calcium-Binding Motif Stabilizes and Increases the Activity of Aspergillus fumigatus Ecto-NADase.
Biochemistry, 62, 2023
|
|
8PMR
 
 | | NADase from Aspergillus fumigatus with mutated calcium binding motif (D219A/E220A) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Ferrario, E, Stromland, O, Ziegler, M. | | Deposit date: | 2023-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel Calcium-Binding Motif Stabilizes and Increases the Activity of Aspergillus fumigatus Ecto-NADase.
Biochemistry, 62, 2023
|
|
6JQE
 
 | | The structural basis of the beta-carbonic anhydrases CafD (wild type) of the filamentous fungus Aspergillus fumigatus | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Jin, M.S, Kim, S, Kim, N.J, Hong, S, Kim, S, Sung, J. | | Deposit date: | 2019-03-30 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of the low catalytic activities of the two minor beta-carbonic anhydrases of the filamentous fungus Aspergillus fumigatus.
J.Struct.Biol., 208, 2019
|
|
6JQC
 
 | | The structural basis of the beta-carbonic anhydrase CafC (wild type) of the filamentous fungus Aspergillus fumigatus | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Jin, M.S, Kim, S, Kim, N.J, Hong, S, Kim, S, Sung, J. | | Deposit date: | 2019-03-30 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of the low catalytic activities of the two minor beta-carbonic anhydrases of the filamentous fungus Aspergillus fumigatus.
J.Struct.Biol., 208, 2019
|
|
6JQD
 
 | | The structural basis of the beta-carbonic anhydrase CafC (L25G and L78G mutant) of the filamentous fungus Aspergillus fumigatus | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Jin, M.S, Kim, S, Kim, N.J, Hong, S, Kim, S, Sung, J. | | Deposit date: | 2019-03-30 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The structural basis of the low catalytic activities of the two minor beta-carbonic anhydrases of the filamentous fungus Aspergillus fumigatus.
J.Struct.Biol., 208, 2019
|
|
4V63
 
 | | Structural basis for translation termination on the 70S ribosome. | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Laurberg, M, Asahara, H, Korostelev, A, Zhu, J, Trakhanov, S, Noller, H.F. | | Deposit date: | 2008-05-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.207 Å) | | Cite: | Structural basis for translation termination on the 70S ribosome
Nature, 454, 2008
|
|
5FOQ
 
 | | Acetylcholinesterase in complex with C7653 | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2,4-dichlorophenoxy)-N-[4-(1-piperidinylmethyl)phenyl]acetamide, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Berg, L, Mishra, B.K, Andersson, D.C, Ekstrom, F, Linusson, A. | | Deposit date: | 2015-11-25 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Nature of Activated Non-Classical Hydrogen Bonds: A Case Study on Acetylcholinesterase-Ligand Complexes.
Chemistry, 22, 2016
|
|
7ZQ9
 
 | | Dimeric PSI of Chlamydomonas reinhardtii at 2.74 A resolution (symmetry expanded) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
7ZQC
 
 | | Monomeric PSI of Chlamydomonas reinhardtii at 2.31 A resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
7ZQE
 
 | |
7ZQD
 
 | | Dimeric PSI of Chlamydomonas reinhardtii at 2.97 A resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
4HZU
 
 | | Structure of a bacterial energy-coupling factor transporter | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA 1, Energy-coupling factor transporter ATP-binding protein EcfA 2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Wang, T.L, Fu, G.B, Pan, X.J, Shi, Y.G. | | Deposit date: | 2012-11-15 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Structure of a bacterial energy-coupling factor transporter.
Nature, 497, 2013
|
|
1WNO
 
 | | Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, ... | | Authors: | Hu, H, Wang, G, Yang, H, Zhou, J, Mo, L, Yang, K, Jin, C, Jin, C, Rao, Z. | | Deposit date: | 2004-08-07 | | Release date: | 2005-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407
To be Published
|
|
1G7I
 
 | | CRYSTAL STRUCTURE OF HEN EGG WHITE LYSOZYME (HEL) COMPLEXED WITH THE MUTANT ANTI-HEL MONOCLONAL ANTIBODY D1.3 (VLW92F) | | Descriptor: | ANTI-HEN EGG WHITE LYSOZYME MONOCLONAL ANTIBODY D1.3, LYSOZYME C, PHOSPHATE ION | | Authors: | Sundberg, E.J, Urrutia, M, Braden, B.C, Isern, J, Mariuzza, R.A. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Estimation of the hydrophobic effect in an antigen-antibody protein-protein interface.
Biochemistry, 39, 2000
|
|
1G7L
 
 | | CRYSTAL STRUCTURE OF HEN EGG WHITE LYSOZYME (HEL) COMPLEXED WITH THE MUTANT ANTI-HEL MONOCLONAL ANTIBODY D1.3 (VLW92S) | | Descriptor: | ANTI-HEN EGG WHITE LYSOZYME MONOCLONAL ANTIBODY D1.3, LYSOZYME C | | Authors: | Sundberg, E.J, Urrutia, M, Braden, B.C, Isern, J, Mariuzza, R.A. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Estimation of the hydrophobic effect in an antigen-antibody protein-protein interface.
Biochemistry, 39, 2000
|
|
1G7M
 
 | | CRYSTAL STRUCTURE OF HEN EGG WHITE LYSOZYME (HEL) COMPLEXED WITH THE MUTANT ANTI-HEL MONOCLONAL ANTIBODY D1.3 (VLW92V) | | Descriptor: | ANTI-HEN EGG WHITE LYSOZYME MONOCLONAL ANTIBODY D1.3, LYSOZYME C | | Authors: | Sundberg, E.J, Urrutia, M, Braden, B.C, Isern, J, Mariuzza, R.A. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Estimation of the hydrophobic effect in an antigen-antibody protein-protein interface.
Biochemistry, 39, 2000
|
|
1KMP
 
 | | Crystal structure of the Outer Membrane Transporter FecA Complexed with Ferric Citrate | | Descriptor: | CITRIC ACID, FE (III) ION, IRON(III) DICITRATE TRANSPORT PROTEIN FECA, ... | | Authors: | Ferguson, A.D, Chakraborty, R, Smith, B.S, Esser, L, van der Helm, D, Deisenhofer, J. | | Deposit date: | 2001-12-17 | | Release date: | 2002-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of gating by the outer membrane transporter FecA.
Science, 295, 2002
|
|
