5CQK
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3B, GLYCEROL, SODIUM ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain.
J.Biol.Chem., 290, 2015
|
|
5CQH
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain.
J.Biol.Chem., 290, 2015
|
|
5CQI
 
 | |
5CQD
 
 | |
1UHG
 
 | | Crystal Structure of S-Ovalbumin At 1.9 Angstrom Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ovalbumin, ... | | Authors: | Yamasaki, M, Takahashi, N, Hirose, M. | | Deposit date: | 2003-07-03 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of S-ovalbumin as a Non-loop-inserted Thermostabilized Serpin Form
J.Biol.Chem., 278, 2003
|
|
5D48
 
 | | Crystal Structure of FABP4 in complex with 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy) phenyl]-1H-indol-1-yl}propanoic acid | | Descriptor: | 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy)phenyl]-1H-indol-1-yl}propanoic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
1TFO
 
 | | Ribonuclease from Escherichia coli complexed with its inhibitor protein | | Descriptor: | Colicin D, Colicin D immunity protein | | Authors: | Yajima, S, Nakanishi, K, Takahashi, K, Ogawa, T, Kezuka, Y, Hidaka, M, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2004-05-27 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relation between tRNase activity and the structure of colicin D according to X-ray crystallography
Biochem.Biophys.Res.Commun., 322, 2004
|
|
5D4A
 
 | | Crystal Structure of FABP4 in complex with 3-(2-phenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(2-phenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
1T0D
 
 | | Crystal Structure of 2-aminopurine labelled bacterial decoding site RNA | | Descriptor: | 5'-R(*CP*AP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*CP*C)-3', 5'-R(*GP*GP*UP*GP*GP*UP*GP*(MTU)P*AP*GP*UP*CP*GP*CP*UP*GP*G)-3' | | Authors: | Shandrick, S, Zhao, Q, Han, Q, Ayida, B.K, Takahashi, M, Winters, G.C, Simonsen, K.B, Vourloumis, D, Hermann, T. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Monitoring molecular recognition of the ribosomal decoding site.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1TFK
 
 | | Ribonuclease from Escherichia coli complexed with its inhibtor protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Colicin D, Colicin D immunity protein | | Authors: | Yajima, S, Nakanishi, K, Takahashi, K, Ogawa, T, Kezuka, Y, Hidaka, M, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2004-05-27 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Relation between tRNase activity and the structure of colicin D according to X-ray crystallography
Biochem.Biophys.Res.Commun., 322, 2004
|
|
1UJ8
 
 | |
5D45
 
 | | Crystal Structure of FABP4 in complex with 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid | | Descriptor: | 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D47
 
 | | Crystal Structure of FABP4 in complex with 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl] propanoic acid | | Descriptor: | 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl]propanoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5AX3
 
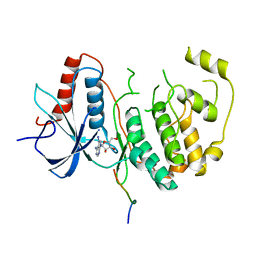 | | Crystal structure of ERK2 complexed with allosteric and ATP-competitive inhibitors. | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Mitogen-activated protein kinase 1, allosteric and ATP-competitive inhibitor | | Authors: | Kinoshita, T, Sugiyama, H, Mori, Y, Takahashi, N, Tomonaga, A. | | Deposit date: | 2015-07-14 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | Identification of allosteric ERK2 inhibitors through in silico biased screening and competitive binding assay
Bioorg.Med.Chem.Lett., 26, 2016
|
|
1T0E
 
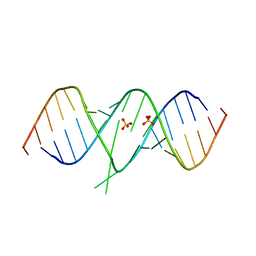 | | Crystal Structure of 2-aminopurine labelled bacterial decoding site RNA | | Descriptor: | 5'-R(*CP*GP*AP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*CP*C)-3', 5'-R(*GP*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*UP*CP*GP*G)-3', SULFATE ION | | Authors: | Shandrick, S, Zhao, Q, Han, Q, Ayida, B.K, Takahashi, M, Winters, G.C, Simonsen, K.B, Vourloumis, D, Hermann, T. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Monitoring molecular recognition of the ribosomal decoding site.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1TAB
 
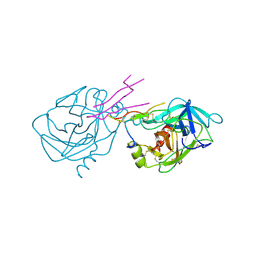 | | STRUCTURE OF THE TRYPSIN-BINDING DOMAIN OF BOWMAN-BIRK TYPE PROTEASE INHIBITOR AND ITS INTERACTION WITH TRYPSIN | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR, TRYPSIN | | Authors: | Tsunogae, Y, Tanaka, I, Yamane, T, Kikkawa, J.-I, Ashida, T, Ishikawa, C, Watanabe, K, Nakamura, S, Takahashi, K. | | Deposit date: | 1990-10-15 | | Release date: | 1992-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the trypsin-binding domain of Bowman-Birk type protease inhibitor and its interaction with trypsin.
J.Biochem.(Tokyo), 100, 1986
|
|
1USC
 
 | |
1VDN
 
 | |
7E5V
 
 | | Crystal structure of Phm7 in complex with inhibitor | | Descriptor: | Diels-Alderase, GLYCEROL, SULFATE ION, ... | | Authors: | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7E5U
 
 | | Crystal structure of Phm7 | | Descriptor: | CHLORIDE ION, Diels-Alderase, GLYCEROL, ... | | Authors: | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7E5T
 
 | | Crystal structure of Fsa2 | | Descriptor: | Diels-Alderase fsa2, ETHANOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.16977525 Å) | | Cite: | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3VHX
 
 | | The crystal structure of Arf6-MKLP1 (Mitotic kinesin-like protein 1) complex | | Descriptor: | ADP-ribosylation factor 6, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Makyio, H, Takei, T, Ohgi, H, Takahashi, S, Takatsu, H, Ueda, T, Kanaho, Y, Xie, Y, Shin, H.W, Kamikubo, H, Kataoka, M, Kawasaki, M, Kato, R, Wakatsuki, S, Nakayama, K. | | Deposit date: | 2011-09-12 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for Arf6-MKLP1 complex formation on the Flemming body responsible for cytokinesis
Embo J., 31, 2012
|
|
3VKP
 
 | | Assimilatory nitrite reductase (Nii3) - NO2 complex from tobbaco leaf analysed with low X-ray dose | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
3VUP
 
 | | Beta-1,4-mannanase from the common sea hare Aplysia kurodai | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-1,4-mannanase, SULFATE ION | | Authors: | Mizutani, K, Tsuchiya, S, Toyoda, M, Nanbu, Y, Tominaga, K, Yuasa, K, Takahashi, N, Tsuji, A, Mikami, B. | | Deposit date: | 2012-07-04 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure of beta-1,4-mannanase from the common sea hare Aplysia kurodai at 1.05 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3VHM
 
 | | Crystal structure of NPC-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4R,6aR)-1-{[(1R)-1-(6-nitro-1,3-benzodioxol-5-yl)ethoxy]carbonyl}-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-29 | | Release date: | 2011-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
