6BGY
 
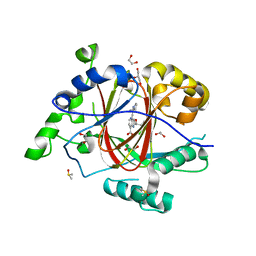 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(1-methylpyrrolidin-2-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound 46) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(S)-(2-chlorophenyl){2-[(2R)-1-methylpyrrolidin-2-yl]ethoxy}methyl]-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6Z9K
 
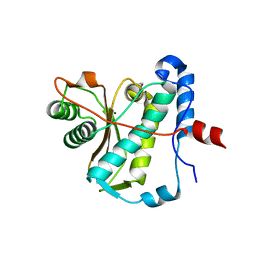 | | CAP domain of Enterococcal PrgA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PrgA | | Authors: | Berntsson, R.P.A, Schmitt, A. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enterococcal PrgA Extends Far Outside the Cell and Provides Surface Exclusion to Protect against Unwanted Conjugation.
J.Mol.Biol., 432, 2020
|
|
6Q3F
 
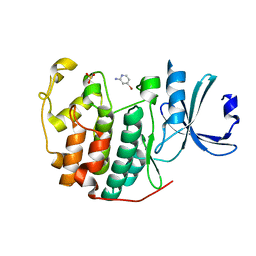 | | CDK2 in complex with FragLite2 | | Descriptor: | 4-bromanylpyridin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6FYE
 
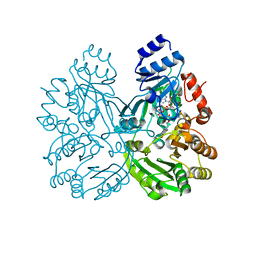 | | The crystal structure of EncM H138T mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Saleem-Batcha, R, Teufel, R. | | Deposit date: | 2018-03-11 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic control of dioxygen binding and functionalization of the flavin cofactor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5IVE
 
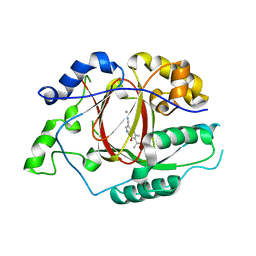 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N8 ( 5-methyl-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile) | | Descriptor: | 5-methyl-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, MANGANESE (II) ION | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
6BGX
 
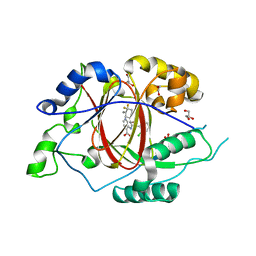 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)((4,4-difluorocyclohexyl)methoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound N42) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[(4,4-difluorocyclohexyl)methoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
8YHF
 
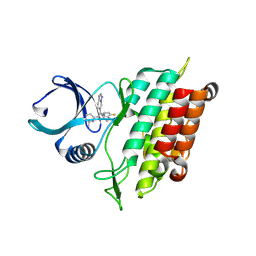 | | The Crystal Structure of the Type I TGF beta receptor from Biortus. | | Descriptor: | 2-fluoranyl-~{N}-[[5-(6-methylpyridin-2-yl)-4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-1~{H}-imidazol-2-yl]methyl]aniline, TGF-beta receptor type-1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Type I TGF beta receptor from Biortus.
To Be Published
|
|
6YU1
 
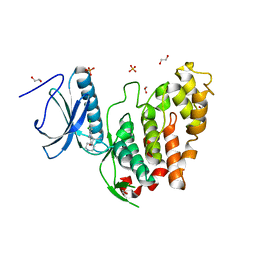 | | CLK3 bound with beta-carboline KH-CARB13 (Cpd 3) | | Descriptor: | (4~{S})-7,8-bis(chloranyl)-9-methyl-1-oxidanylidene-spiro[2,4-dihydropyrido[3,4-b]indole-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3, ... | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6QHM
 
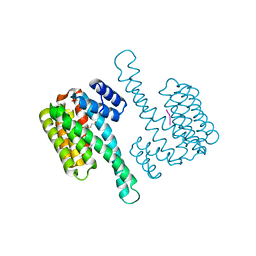 | | 14-3-3 sigma with RelA/p65 binding site pS281 | | Descriptor: | 14-3-3 protein sigma, LEU-SEP-GLU | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2019-01-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Selectivity via Cooperativity: Preferential Stabilization of the p65/14-3-3 Interaction with Semisynthetic Natural Products.
J.Am.Chem.Soc., 142, 2020
|
|
6QK0
 
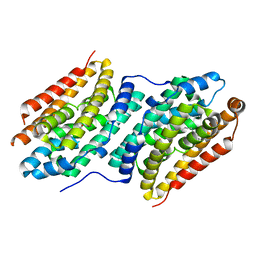 | |
5LA1
 
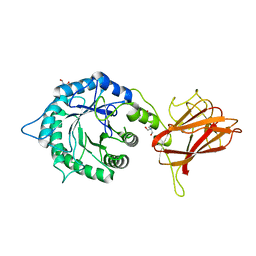 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, H. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
6ZA0
 
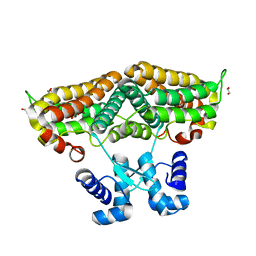 | | Structure of the transcriptional repressor Atu1419 (VanR) in complex with a fortuitous citrate from agrobacterium fabrum (P21212 space group) | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
6ZA3
 
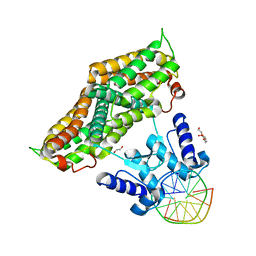 | | Structure of the transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum in complex a palindromic DNA (C2221 space group) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
6YVZ
 
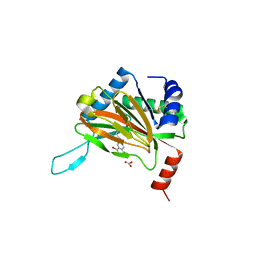 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with bicyclic JLS-367 | | Descriptor: | 4-[(5-bromanylisoquinolin-3-yl)amino]-4-oxidanylidene-butanoic acid, BICARBONATE ION, Egl nine homolog 1, ... | | Authors: | Chowdhury, R, Sorensen, J.L, Schofield, C.J. | | Deposit date: | 2020-04-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Use of cyclic peptides to induce crystallization: case study with prolyl hydroxylase domain 2.
Sci Rep, 10, 2020
|
|
6ZA4
 
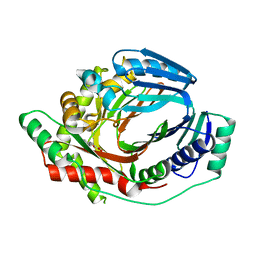 | | M. tuberculosis salicylate synthase MbtI in complex with 5-(3-cyanophenyl)furan-2-carboxylate | | Descriptor: | 5-(3-cyanophenyl)furan-2-carboxylic acid, CHLORIDE ION, Salicylate synthase | | Authors: | Mori, M, Villa, S, Meneghetti, F, Bellinzoni, M. | | Deposit date: | 2020-06-04 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Shedding X-ray Light on the Role of Magnesium in the Activity ofMycobacterium tuberculosisSalicylate Synthase (MbtI) for Drug Design.
J.Med.Chem., 63, 2020
|
|
6GEI
 
 | |
8XQK
 
 | | The Crystal Structure of Apaf from Biortus. | | Descriptor: | Apoptotic protease-activating factor 1, PHOSPHATE ION | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Ni, C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Crystal Structure of Apaf from Biortus.
To Be Published
|
|
8YHP
 
 | | Structure of the PGK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Phosphoglycerate kinase 1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the PGK1 from Biortus.
To Be Published
|
|
6YYH
 
 | | Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum in ligand-free form | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase, CITRIC ACID, ... | | Authors: | Lafite, P, Daniellou, R, Bretagne, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of Dictyoglomus thermophilum beta-d-xylosidase DtXyl unravels the structural determinants for efficient notoginsenoside R1 hydrolysis.
Biochimie, 181, 2020
|
|
6QLG
 
 | | Crystal structure of AnUbiX (PadA1) in complex with FMN and dimethylallyl pyrophosphate | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYLALLYL DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Marshall, S.A, Payne, K.A.P, Leys, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The UbiX flavin prenyltransferase reaction mechanism resembles class I terpene cyclase chemistry.
Nat Commun, 10, 2019
|
|
6FNR
 
 | | Ergothioneine-biosynthetic methyltransferase EgtD in complex with chlorohistidine | | Descriptor: | (2~{S})-2-chloranyl-3-(1~{H}-imidazol-5-yl)propanoic acid, GLYCEROL, Histidine N-alpha-methyltransferase, ... | | Authors: | Vit, A, Blankenfeldt, W, Seebeck, F.P. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Inhibition and Regulation of the Ergothioneine Biosynthetic Methyltransferase EgtD.
ACS Chem. Biol., 13, 2018
|
|
6G0K
 
 | | Crystal structure of Enterococcus faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | Descriptor: | Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | Authors: | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2018-03-19 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
8Y9A
 
 | |
6BWU
 
 | | Crystal structure of carboxyhemoglobin in complex with beta Cys93 modifying agent, TD3 | | Descriptor: | 1H-1,2,3-triazole-5-thiol, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Musayev, F.N, Safo, R.M, Safo, M.K. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Triazole Disulfide Compound Increases the Affinity of Hemoglobin for Oxygen and Reduces the Sickling of Human Sickle Cells.
Mol. Pharm., 15, 2018
|
|
8AF1
 
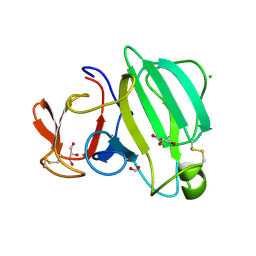 | | Beta-Lytic Protease from Lysobacter capsici | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kudryakova, I.V, Afoshin, A.S, Vasilyeva, N.V. | | Deposit date: | 2022-07-15 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural and Functional Characterization of beta-lytic Protease from Lysobacter capsici VKM B-2533 T.
Int J Mol Sci, 23, 2022
|
|
