2H8K
 
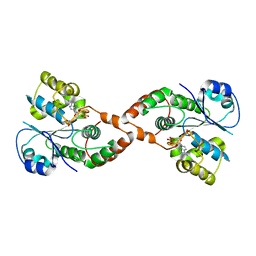 | | Human Sulfotranferase SULT1C3 in complex with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, SULT1C3 splice variant d | | Authors: | Tempel, W, Dombrovski, L, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-07 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and chemical profiling of the human cytosolic sulfotransferases.
Plos Biol., 5, 2007
|
|
2H8L
 
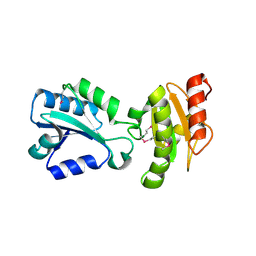 | |
2H8M
 
 | | N-Domain Of Grp94 In Complex With the 2-Iodo-NECA | | Descriptor: | (2S,3S,4R,5R)-5-(6-AMINO-2-IODO-9H-PURIN-9-YL)-N-ETHYL-3,4-DIHYDROXYTETRAHYDROFURAN-2-CARBOXAMIDE, Endoplasmin, PENTAETHYLENE GLYCOL, ... | | Authors: | Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2006-06-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N-Domain Of Grp94 In Complex With the 2-Iodo-NECA
To be Published
|
|
2H8N
 
 | |
2H8O
 
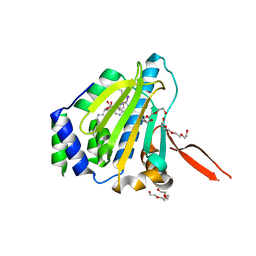
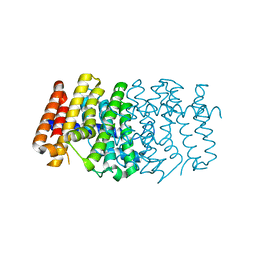 | | The 1.6A crystal structure of the geranyltransferase from Agrobacterium tumefaciens | | Descriptor: | Geranyltranstransferase | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-06-07 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6A crystal structure of the geranyltransferase from Agrobacterium tumefaciens
To be Published
|
|
2H8P
 
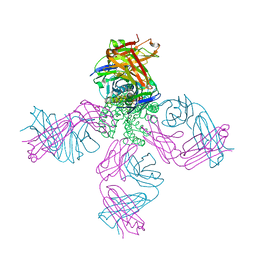 | | Structure of a K channel with an amide to ester substitution in the selectivity filter | | Descriptor: | (2S)-2-(BUTYRYLOXY)-3-HYDROXYPROPYL NONANOATE, FAB heavy chain, FAB light chain, ... | | Authors: | Valiyaveetil, F.I, MacKinnon, R, Muir, T.W. | | Deposit date: | 2006-06-07 | | Release date: | 2006-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and Functional Consequences of an Amide-to-Ester Substitution in the Selectivity Filter of a Potassium Channel.
J.Am.Chem.Soc., 128, 2006
|
|
2H8Q
 
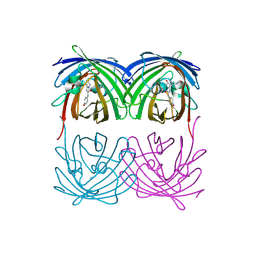 | |
2H8R
 
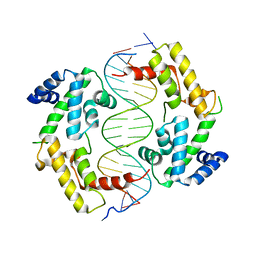 | | Hepatocyte Nuclear Factor 1b bound to DNA: MODY5 Gene Product | | Descriptor: | 5'-D(*CP*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*TP*CP*AP*CP*CP*AP*G)-3', 5'-D(*GP*CP*TP*GP*GP*TP*GP*AP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*A)-3', Hepatocyte nuclear factor 1-beta | | Authors: | Lu, P, Rha, G.B, Chi, Y.I. | | Deposit date: | 2006-06-07 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of disease-causing mutations in hepatocyte nuclear factor 1beta.
Biochemistry, 46, 2007
|
|
2H8S
 
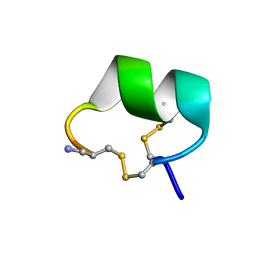 | | Solution structure of alpha-conotoxin Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Clark, R.J, Fischer, H, Nevin, S.T, Adams, D.J, Craik, D.J. | | Deposit date: | 2006-06-07 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | The Synthesis, Structural Characterization, and Receptor Specificity of the {alpha}-Conotoxin Vc1.1.
J.Biol.Chem., 281, 2006
|
|
2H8U
 
 | |
2H8V
 
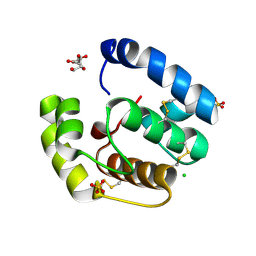 | | Structure of empty Pheromone Binding Protein ASP1 from the Honeybee Apis mellifera L | | Descriptor: | CHLORIDE ION, GLYCEROL, Pheromone-binding protein ASP1, ... | | Authors: | Pesenti, M.E, Spinelli, S, Briand, L, Pernollet, J.-C, Cambillau, C, Tegoni, M. | | Deposit date: | 2006-06-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational Changes of the Pheromone Binding Protein ASP1 from the Honeybee Apis mellifera L upon Ligand Binding
To be Published
|
|
2H8W
 
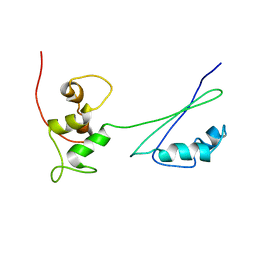 | | Solution structure of ribosomal protein L11 | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Choli-Papadopoulou, T, Krueger, S, Draper, D, Wang, Y.-X. | | Deposit date: | 2006-06-08 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of Free L11 and Functional Dynamics of L11 in Free, L11-rRNA(58 nt) Binary and L11-rRNA(58 nt)-thiostrepton Ternary Complexes.
J.Mol.Biol., 367, 2007
|
|
2H8X
 
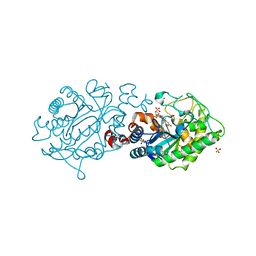 | | Xenobiotic Reductase A-oxidized | | Descriptor: | FLAVIN MONONUCLEOTIDE, SULFATE ION, Xenobiotic reductase A | | Authors: | Dobbek, H. | | Deposit date: | 2006-06-08 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Xenobiotic reductase A in the degradation of quinoline by Pseudomonas putida 86: physiological function, structure and mechanism of 8-hydroxycoumarin reduction.
J.Mol.Biol., 361, 2006
|
|
2H8Z
 
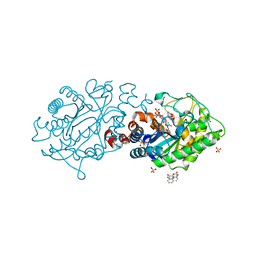 | | Xenobiotic Reductase A in complex with 8-Hydroxycoumarin | | Descriptor: | 8-HYDROXYCOUMARIN, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Dobbek, H. | | Deposit date: | 2006-06-08 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Xenobiotic reductase A in the degradation of quinoline by Pseudomonas putida 86: physiological function, structure and mechanism of 8-hydroxycoumarin reduction.
J.Mol.Biol., 361, 2006
|
|
2H90
 
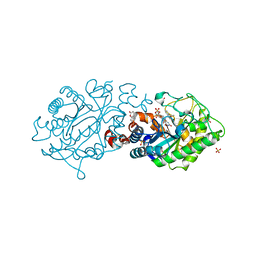 | | Xenobiotic reductase A in complex with coumarin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, COUMARIN, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobbek, H. | | Deposit date: | 2006-06-08 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Xenobiotic reductase A in the degradation of quinoline by Pseudomonas putida 86: physiological function, structure and mechanism of 8-hydroxycoumarin reduction.
J.Mol.Biol., 361, 2006
|
|
2H92
 
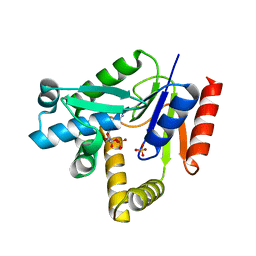 | |
2H94
 
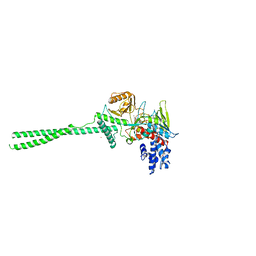 | |
2H95
 
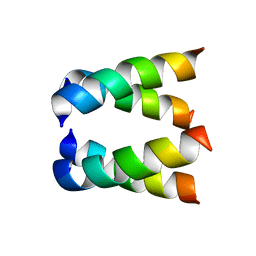 | |
2H96
 
 | | Discovery of Potent, Highly Selective, and Orally Bioavailable Pyridine Carboxamide C-jun NH2-terminal Kinase Inhibitors | | Descriptor: | 5-CYANO-N-(2,5-DIMETHOXYBENZYL)-6-ETHOXYPYRIDINE-2-CARBOXAMIDE, C-jun-amino-terminal kinase-interacting protein 1, GLYCEROL, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-06-09 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of potent, highly selective, and orally bioavailable pyridine carboxamide c-Jun NH2-terminal kinase inhibitors.
J.Med.Chem., 49, 2006
|
|
2H98
 
 | |
2H99
 
 | | Crystal structure of the effector binding domain of a BenM variant (R156H,T157S) | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ezezika, O.C, Craven, S.H, Neidle, E.L, Momany, C. | | Deposit date: | 2006-06-09 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Inducer responses of BenM, a LysR-type transcriptional regulator from Acinetobacter baylyi ADP1.
Mol.Microbiol., 72, 2009
|
|
2H9A
 
 | | Corrinoid Iron-Sulfur Protein | | Descriptor: | CO dehydrogenase/acetyl-CoA synthase, iron-sulfur protein, COBALAMIN, ... | | Authors: | Dobbek, H. | | Deposit date: | 2006-06-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into methyltransfer reactions of a corrinoid iron-sulfur protein involved in acetyl-CoA synthesis.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2H9B
 
 | | Crystal structure of the effector binding domain of a BenM variant (BenM R156H/T157S) | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator benM, SULFATE ION | | Authors: | Ezezika, O.C, Craven, S.H, Neidle, E.L, Momany, C. | | Deposit date: | 2006-06-09 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inducer responses of BenM, a LysR-type transcriptional regulator from Acinetobacter baylyi ADP1.
Mol.Microbiol., 72, 2009
|
|
2H9C
 
 | |
2H9D
 
 | |
