8B8F
 
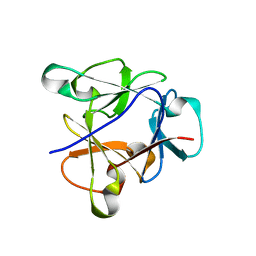 | | Atomic structure of the beta-trefoil domain of the Laccaria bicolor lectin LBL in complex with lactose | | Descriptor: | N-terminal beta-trefoil domain of the lectin LBL from Laccaria bicolor, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Acebron, I, Campanero-Rhodes, M.A, Solis, D, Menendez, M, Garcia, C, Lillo, M.P, Mancheno, J.M. | | Deposit date: | 2022-10-04 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic crystal structure and sugar specificity of a beta-trefoil lectin domain from the ectomycorrhizal basidiomycete Laccaria bicolor.
Int.J.Biol.Macromol., 233, 2023
|
|
8B97
 
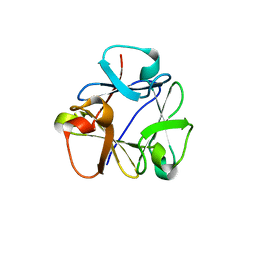 | | N-terminal beta-trefoil lectin domain of the Laccaria bicolor lectin in complex with N-acetyl-lactosamine | | Descriptor: | Beta-trefoil domain of the LBL lectin, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Acebron, I, Campanero-Rhodes, M.A, Solis, D, Menendez, M, Garcia, C, Lillo, M.P, Mancheno, J.M. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Atomic crystal structure and sugar specificity of a beta-trefoil lectin domain from the ectomycorrhizal basidiomycete Laccaria bicolor.
Int.J.Biol.Macromol., 233, 2023
|
|
7OB2
 
 | | NMR structure of the antimicrobial RiLK1 peptide in SDS micelles | | Descriptor: | RiLK1 | | Authors: | Falcigno, L, D'Auria, G, Palmieri, G, Gogliettino, M, Agrillo, B. | | Deposit date: | 2021-04-20 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Key Physicochemical Determinants in the Antimicrobial Peptide RiLK1 Promote Amphipathic Structures.
Int J Mol Sci, 22, 2021
|
|
6OEF
 
 | | PolyAla Model of the O-layer from the Type 4 Secretion System of H. pylori | | Descriptor: | PolyAla Model of OMCC O-Layer | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6OEE
 
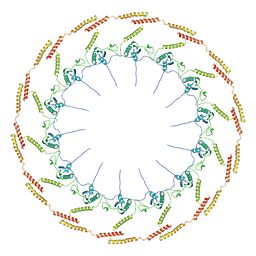 | | Structure of CagT from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagT | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6ODI
 
 | | Structure of CagY from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagY | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
8CYS
 
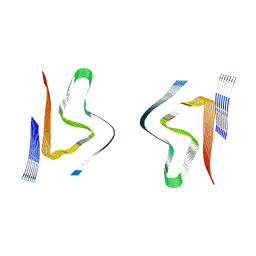 | | CryoEM structures of amplified alpha-synuclein fibril class B type II with extended core from DLB case I | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CZ2
 
 | | CryoEM structure of amplified alpha-synuclein fibril class A type I with extended core from DLB case VII | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CYV
 
 | | CryoEM structures of amplified alpha-synuclein fibril class B mixed type I/II with extended core from DLB case II | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CZ6
 
 | | CryoEM structure of amplified alpha-synuclein fibril class A type I with extended core from DLB case X | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CYX
 
 | | CryoEM structure of amplified alpha-synuclein fibril class A type I with compact core from DLB case III | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CYY
 
 | | CryoEM structure of amplified alpha-synuclein fibril class B mixed type I/II with extended core from DLB case V | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CZ0
 
 | | CryoEM structure of amplified alpha-synuclein fibril class B type I with extended core from DLB case VII | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CYW
 
 | | CryoEM structures of amplified alpha-synuclein fibril class B type I with compact core from DLB case III | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CYT
 
 | | CryoEM structures of amplified alpha-synuclein fibril class A type I with extended core from DLB case I | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CZ1
 
 | | CryoEM structure of amplified alpha-synuclein fibril class B type II with extended core from DLB case VII | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CZ3
 
 | | CryoEM structure of amplified alpha-synuclein fibril class B type I with extended core from DLB case X | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
6OEG
 
 | | Structure of CagX from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagX | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6OEH
 
 | | PolyAla Model of OMCC I-Layer | | Descriptor: | PolyAla Model of OMCC I-Layer | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
3DP4
 
 | | Crystal structure of the binding domain of the AMPA subunit GluR3 bound to AMPA | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Wang, Q, Sondermann, H, Oswald, R.E. | | Deposit date: | 2008-07-07 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of the S1S2 glutamate binding domain of GLuR3.
Proteins, 75, 2008
|
|
8PI0
 
 | |
8PIY
 
 | |
4CNU
 
 | |
4U63
 
 | | Crystal structure of a bacterial class III photolyase from Agrobacterium tumefaciens at 1.67A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA photolyase, ... | | Authors: | Scheerer, P, Zhang, F, Kalms, J, von Stetten, D, Krauss, N, Oberpichler, I, Lamparter, T. | | Deposit date: | 2014-07-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Class III Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals a New Antenna Chromophore Binding Site and Alternative Photoreduction Pathways.
J.Biol.Chem., 290, 2015
|
|
4CNS
 
 | | Crystal structure of truncated human CRMP-4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Crmp-4: Correction of Intensities for Lattice-Translocation Disorder
Acta Crystallogr.,Sect.D, 70, 2014
|
|
