3WEU
 
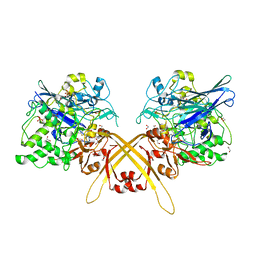 | | Crystal structure of the L-Lys epsilon-oxidase from Marinomonas mediterranea | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, L-lysine 6-oxidase, ... | | Authors: | Okazaki, S, Nakano, S, Matsui, D, Akaji, S, Inagaki, K, Asano, Y. | | Deposit date: | 2013-07-12 | | Release date: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-Ray crystallographic evidence for the presence of the cysteine tryptophylquinone cofactor in L-lysine {varepsilon}-oxidase from Marinomonas mediterranea
J.Biochem., 154, 2013
|
|
5OQM
 
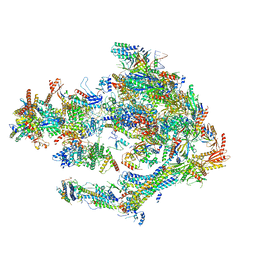 | | STRUCTURE OF YEAST TRANSCRIPTION PRE-INITIATION COMPLEX WITH TFIIH AND CORE MEDIATOR | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Schilbach, S, Hantsche, M, Tegunov, D, Dienemann, C, Wigge, C, Urlaub, H, Cramer, P. | | Deposit date: | 2017-08-13 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of transcription pre-initiation complex with TFIIH and Mediator.
Nature, 551, 2017
|
|
4BIK
 
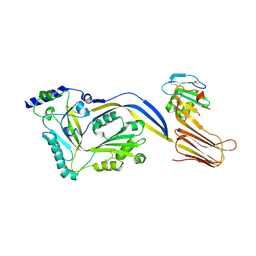 | | Structure of a disulfide locked mutant of Intermedilysin with human CD59 | | Descriptor: | CD59 GLYCOPROTEIN, INTERMEDILYSIN | | Authors: | Johnson, S, Brooks, N.J, Smith, R.A.G, Lea, S.M, Bubeck, D. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.494 Å) | | Cite: | Structural Basis for Recognition of the Pore- Forming Toxin Intermedilysin by Human Complement Receptor Cd59
Cell Rep., 3, 2013
|
|
1RCJ
 
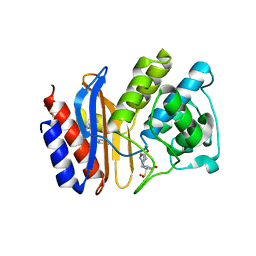 | | Crystal structure of E166A mutant of SHV-1 beta-lactamase with the trans-enamine intermediate of tazobactam | | Descriptor: | Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, TAZOBACTAM TRANS-ENAMINE INTERMEDIATE | | Authors: | Padayatti, P.S, Helfand, M.S, Totir, M.A, Carey, M.P, Hujer, A.M, Carey, P.R, Bonomo, R.A, van den Akker, F. | | Deposit date: | 2003-11-04 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Tazobactam Forms a Stoichiometric trans-Enamine Intermediate in the E166A Variant of SHV-1 beta-Lactamase: 1.63 A Crystal Structure
Biochemistry, 43, 2004
|
|
2LJY
 
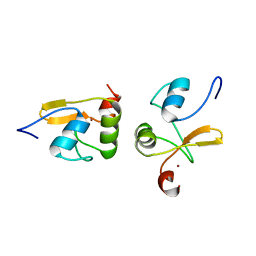 | | Haddock model structure of the N-terminal domain dimer of HPV16 E6 | | Descriptor: | Protein E6, ZINC ION | | Authors: | Zanier, K, Muhamed Sidi, A, Boulade-Ladame, C, Rybin, V, Chappelle, A, Atkinson, A, Kieffer, B, Trave, G. | | Deposit date: | 2011-09-30 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Analysis of the HPV16 E6 Oncoprotein Reveals a Self-Association Mechanism Required for E6-Mediated Degradation of p53.
Structure, 20, 2012
|
|
2LJX
 
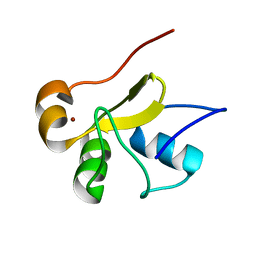 | | Structure of the monomeric N-terminal domain of HPV16 E6 oncoprotein | | Descriptor: | Protein E6, ZINC ION | | Authors: | Zanier, K, Muhamed Sidi, A, Boulade-Ladame, C, Rybin, V, Chappelle, A, Atkinson, A, Kieffer, B, Trave, G. | | Deposit date: | 2011-09-30 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Analysis of the HPV16 E6 Oncoprotein Reveals a Self-Association Mechanism Required for E6-Mediated Degradation of p53.
Structure, 20, 2012
|
|
2LJZ
 
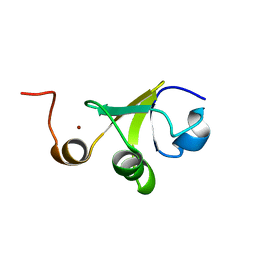 | | Structure of the C-terminal domain of HPV16 E6 oncoprotein | | Descriptor: | Protein E6, ZINC ION | | Authors: | Zanier, K, Muhamed Sidi, A, Boulade-Ladame, C, Rybin, V, Chappelle, A, Atkinson, A, Kieffer, B, Trave, G. | | Deposit date: | 2011-09-30 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Analysis of the HPV16 E6 Oncoprotein Reveals a Self-Association Mechanism Required for E6-Mediated Degradation of p53.
Structure, 20, 2012
|
|
5WX8
 
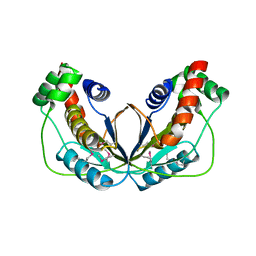 | | Human herpesvirus 6A immediate early protein 2 C-terminal domain | | Descriptor: | Immediate-early protein 2 | | Authors: | Nishimura, M, Wang, J, Wakata, A, Sakamoto, K, Mori, Y. | | Deposit date: | 2017-01-06 | | Release date: | 2017-08-02 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the DNA-Binding Domain of Human Herpesvirus 6A Immediate Early Protein 2.
J. Virol., 91, 2017
|
|
8J9A
 
 | |
2GAS
 
 | | Crystal Structure of Isoflavone Reductase | | Descriptor: | isoflavone reductase | | Authors: | Wang, X, He, X, Lin, J, Shao, H, Chang, Z, Dixon, R.A. | | Deposit date: | 2006-03-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Isoflavone Reductase from Alfalfa (Medicago sativa L.)
J.Mol.Biol., 358, 2006
|
|
6VCW
 
 | | Crystal structure of Medicago truncatula S-adenosylmethionine Synthase 3A (MtMAT3A) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
5EQ8
 
 | |
5EQA
 
 | |
5EQ7
 
 | |
5EQ9
 
 | |
2FLY
 
 | | Proadrenomedullin N-Terminal 20 Peptide | | Descriptor: | Proadrenomedullin N-20 terminal peptide | | Authors: | Lucyk, S, Taha, H, Yamamoto, H, Miskolzie, M, Kotovych, G. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR conformational analysis of proadrenomedullin N-terminal 20 peptide, a proangiogenic factor involved in tumor growth
Biopolymers, 81, 2006
|
|
5BSH
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) in complex with L-Proline | | Descriptor: | PROLINE, Pyrroline-5-carboxylate reductase | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
7CKE
 
 | |
7CKD
 
 | |
1ZB8
 
 | | Crystal structure of Xylella fastidiosa organic peroxide resistance protein | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, organic hydroperoxide resistance protein | | Authors: | Oliveira, M.A, Guimaraes, B.G, Cussiol, J.R, Medrano, F.J, Vidigal, S.A, Gozzo, F.C, Netto, L.E. | | Deposit date: | 2005-04-07 | | Release date: | 2006-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into Enzyme-Substrate Interaction and Characterization of Enzymatic Intermediates of Organic Hydroperoxide Resistance Protein from Xylella fastidiosa.
J.Mol.Biol., 359, 2006
|
|
5BSF
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) in complex with NAD+ | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
1ZB9
 
 | | Crystal structure of Xylella fastidiosa organic peroxide resistance protein | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, organic hydroperoxide resistance protein | | Authors: | Oliveira, M.A, Guimaraes, B.G, Cussiol, J.R, Medrano, F.J, Vidigal, S.A, Gozzo, F.C, Netto, L.E. | | Deposit date: | 2005-04-07 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into Enzyme-Substrate Interaction and Characterization of Enzymatic Intermediates of Organic Hydroperoxide Resistance Protein from Xylella fastidiosa.
J.Mol.Biol., 359, 2006
|
|
5BSE
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, Pyrroline-5-carboxylate reductase | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
5BSG
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) in complex with NADP+ | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
4B4D
 
 | | Crystal structure of FAD-containing ferredoxin-NADP reductase from Xanthomonas axonopodis pv. citri | | Descriptor: | CHLORIDE ION, FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Martinez-Julvez, M, Orellano, E.G, Tondo, M.L, Hurtado-Guerrero, R, Medina, M, Ceccarelli, E.A, Sanchez-Azqueta, A. | | Deposit date: | 2012-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Fad-Containing Ferredoxin- Nadp Reductase from Xanthomonas Axonopodis Pv. Citri
Biomed Res Int, 2013, 2013
|
|
