2FU3
 
 | |
2FU4
 
 | | Crystal Structure of the DNA binding domain of E.coli FUR (Ferric Uptake Regulator) | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferric uptake regulation protein, ... | | Authors: | Pecqueur, L, D'Autreaux, B, Dupuy, J, Nicolet, Y, Jacquamet, L, Brutscher, B, Michaud-Soret, I, Bersch, B. | | Deposit date: | 2006-01-26 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural changes of Escherichia coli ferric uptake regulator during metal-dependent dimerization and activation explored by NMR and X-ray crystallography
J.Biol.Chem., 281, 2006
|
|
2FU5
 
 | | structure of Rab8 in complex with MSS4 | | Descriptor: | BETA-MERCAPTOETHANOL, Guanine nucleotide exchange factor MSS4, Ras-related protein Rab-8A, ... | | Authors: | Itzen, A, Pylypenko, O, Goody, R.S, Rak, A. | | Deposit date: | 2006-01-26 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide exchange via local protein unfolding-structure of Rab8 in complex with MSS4
Embo J., 25, 2006
|
|
2FU6
 
 | |
2FU7
 
 | | Zinc-beta-lactamase L1 from stenotrophomonas maltophilia (Cu-substituted form) | | Descriptor: | 1,10-PHENANTHROLINE, COPPER (II) ION, GLYCEROL, ... | | Authors: | Nauton, L, Garau, G, Kahn, R, Dideberg, O. | | Deposit date: | 2006-01-26 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia.
J.Mol.Biol., 375, 2008
|
|
2FU8
 
 | | Zinc-beta-lactamase L1 from stenotrophomonas maltophilia (d-captopril complex) | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, GLYCEROL, Metallo-beta-lactamase L1, ... | | Authors: | Nauton, L, Garau, G, Kahn, R, Dideberg, O. | | Deposit date: | 2006-01-26 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia.
J.Mol.Biol., 375, 2008
|
|
2FU9
 
 | | Zinc-beta-lactamase L1 from stenotrophomonas maltophilia (mp2 inhibitor complex) | | Descriptor: | GLYCEROL, Metallo-beta-lactamase L1, N-[(BENZYLOXY)CARBONYL]-L-CYSTEINYLGLYCINE, ... | | Authors: | Nauton, L, Garau, G, Kahn, R, Dideberg, O. | | Deposit date: | 2006-01-26 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia.
J.Mol.Biol., 375, 2008
|
|
2FUA
 
 | | L-FUCULOSE 1-PHOSPHATE ALDOLASE CRYSTAL FORM T WITH COBALT | | Descriptor: | BETA-MERCAPTOETHANOL, COBALT (II) ION, L-FUCULOSE-1-PHOSPHATE ALDOLASE, ... | | Authors: | Dreyer, M.K, Schulz, G.E. | | Deposit date: | 1996-02-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined high-resolution structure of the metal-ion dependent L-fuculose-1-phosphate aldolase (class II) from Escherichia coli.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2FUB
 
 | | Crystal structure of urate oxidase at 140 MPa | | Descriptor: | 8-AZAXANTHINE, CYSTEINE, Uricase | | Authors: | Colloc'h, N, Girard, E, Fourme, R. | | Deposit date: | 2006-01-26 | | Release date: | 2006-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High pressure macromolecular crystallography: The 140-MPa crystal structure at 2.3 A resolution of urate oxidase, a 135-kDa tetrameric assembly
Biochim.Biophys.Acta, 1764, 2006
|
|
2FUC
 
 | | Human alpha-Phosphomannomutase 1 with Mg2+ cofactor bound | | Descriptor: | MAGNESIUM ION, Phosphomannomutase 1 | | Authors: | Silvaggi, N.R, Zhang, C, Lu, Z, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structures of human alpha-phosphomannomutase 1 reveal the structural basis of congenital disorder of glycosylation type 1a.
J.Biol.Chem., 281, 2006
|
|
2FUD
 
 | | Human Cathepsin S with Inhibitor CRA-27566 | | Descriptor: | N-{(1R)-2-[(4-CYANO-1,1-DIOXIDOTETRAHYDRO-2H-THIOPYRAN-4-YL)AMINO]-2-OXO-1-[(TRIMETHYLSILYL)METHYL]ETHYL}MORPHOLINE-4-CARBOXAMIDE, cathepsin S | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-26 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human Cathepsin S with Inhibitor CRA-27566
To be Published
|
|
2FUE
 
 | | Human alpha-Phosphomannomutase 1 with D-mannose 1-phosphate and Mg2+ cofactor bound | | Descriptor: | 1-O-phosphono-alpha-D-mannopyranose, MAGNESIUM ION, Phosphomannomutase 1 | | Authors: | Silvaggi, N.R, Zhang, C, Lu, Z, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray crystal structures of human alpha-phosphomannomutase 1 reveal the structural basis of congenital disorder of glycosylation type 1a.
J.Biol.Chem., 281, 2006
|
|
2FUF
 
 | |
2FUG
 
 | |
2FUH
 
 | | Solution Structure of the UbcH5c/Ub Non-covalent Complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Brzovic, P.S, Lissounov, A, Hoyt, D.W, Klevit, R.E. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A UbcH5/Ubiquitin Noncovalent Complex Is Required for Processive BRCA1-Directed Ubiquitination.
Mol.Cell, 21, 2006
|
|
2FUI
 
 | |
2FUJ
 
 | |
2FUK
 
 | | Crystal structure of XC6422 from Xanthomonas campestris: a member of a/b serine hydrolase without lid at 1.6 resolution | | Descriptor: | XC6422 protein | | Authors: | Yang, C.Y, Chin, K.H, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2006-01-27 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of XC6422 from Xanthomonas campestris at 1.6 A resolution: a small serine alpha/beta-hydrolase
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2FUL
 
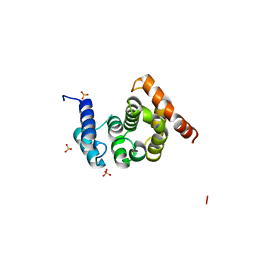 | | Crystal Structure of the C-terminal Domain of S. cerevisiae eIF5 | | Descriptor: | Eukaryotic translation initiation factor 5, SULFATE ION | | Authors: | Wei, Z, Xue, Y, Xu, H, Gong, W. | | Deposit date: | 2006-01-27 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the C-terminal Domain of S.cerevisiae eIF5
J.Mol.Biol., 359, 2006
|
|
2FUM
 
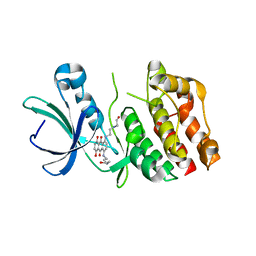 | | Catalytic domain of protein kinase PknB from Mycobacterium tuberculosis in complex with mitoxantrone | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, Probable serine/threonine-protein kinase pknB | | Authors: | Wehenkel, A, Alzari, P.M. | | Deposit date: | 2006-01-27 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The structure of PknB in complex with mitoxantrone, an ATP-competitive inhibitor, suggests a mode of protein kinase regulation in mycobacteria
Febs Lett., 580, 2006
|
|
2FUN
 
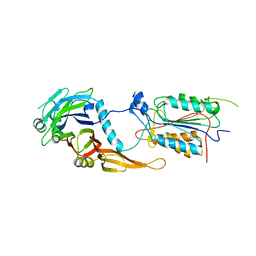 | | alternative p35-caspase-8 complex | | Descriptor: | Early 35 kDa protein, caspase-8 | | Authors: | Lu, M, Min, T, Eliezer, D, Wu, H. | | Deposit date: | 2006-01-27 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Native chemical ligation in covalent caspase inhibition by p35.
Chem.Biol., 13, 2006
|
|
2FUP
 
 | |
2FUQ
 
 | | Crystal Structure of Heparinase II | | Descriptor: | FORMIC ACID, PHOSPHATE ION, ZINC ION, ... | | Authors: | Shaya, D, Cygler, M. | | Deposit date: | 2006-01-27 | | Release date: | 2006-04-18 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Heparinase II from Pedobacter heparinus and Its Complex with a Disaccharide Product.
J.Biol.Chem., 281, 2006
|
|
2FUR
 
 | |
2FUS
 
 | |
