1MVY
 
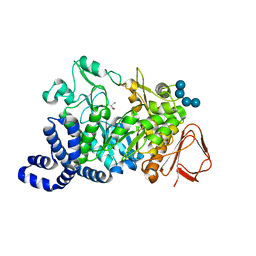 | | Amylosucrase mutant E328Q co-crystallized with maltoheptaose. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, Dar, I, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2002-09-27 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide and Sucrose Complexes of Amylosucrase. STRUCTURAL IMPLICATIONS FOR THE POLYMERASE ACTIVITY
J.BIOL.CHEM., 277, 2002
|
|
1GW6
 
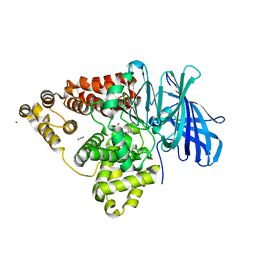 | | STRUCTURE OF LEUKOTRIENE A4 HYDROLASE D375N MUTANT | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, ACETATE ION, IMIDAZOLE, ... | | Authors: | Rudberg, P.C, Tholander, F, Thunnissen, M.M.G.M, Samuelsson, B, Haeggstrom, J.Z. | | Deposit date: | 2002-03-07 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Leukotriene A4 Hydrolase: Selective Abrogation of Leukotriene B4 Formation by Mutation of Aspartic Acid 375
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8B4O
 
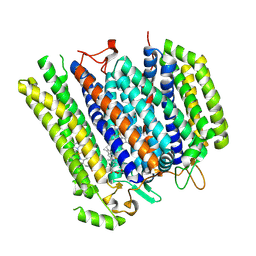 | | Cryo-EM structure of cytochrome bd oxidase from C. glutamicum | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome BD ubiquinol oxidase subunit I, Cytochrome bd-type quinol oxidase subunit II, ... | | Authors: | Grund, T.N, Kusumoto, T, Michel, H, Sakamoto, J, Safarian, S. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Cryo-EM structure of cytochrome bd oxidase from C. glutamicum
To be published
|
|
8BEL
 
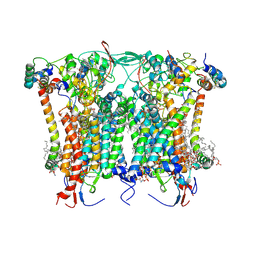 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CIII membrane domain) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BEH
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane tip) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BEF
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane core) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
1R7A
 
 | | Sucrose Phosphorylase from Bifidobacterium adolescentis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, sucrose phosphorylase | | Authors: | Sprogoe, D, van den Broek, L.A.M, Mirza, O, Kastrup, J.S, Voragen, A.G.J, Gajhede, M, Skov, L.K. | | Deposit date: | 2003-10-21 | | Release date: | 2004-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of sucrose phosphorylase from Bifidobacterium adolescentis.
Biochemistry, 43, 2004
|
|
8BEP
 
 | |
8BC7
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex an aminoglutarimide degron peptide | | Descriptor: | Cereblon isoform 4, PHE-PHE-GLU-GLN-MET-GLN-QCI, S-Thalidomide, ... | | Authors: | Heim, C, Albrecht, R, Spring, A.K, Hartmann, M.D. | | Deposit date: | 2022-10-15 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Identification and structural basis of C-terminal cyclic imides as natural degrons for cereblon.
Biochem.Biophys.Res.Commun., 637, 2022
|
|
8BEE
 
 | |
8BED
 
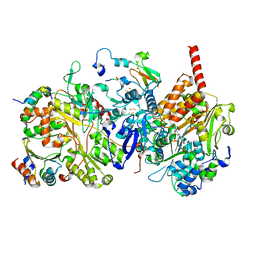 | |
1NL7
 
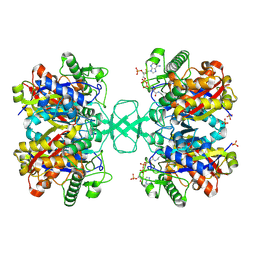 | |
1I9V
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A TRNA-NEOMYCIN COMPLEX | | Descriptor: | MAGNESIUM ION, NEOMYCIN, PHENYLALANINE TRANSFER RNA | | Authors: | Mikkelsen, N.E, Johansson, K, Virtanen, A, Kirsebom, L.A. | | Deposit date: | 2001-03-21 | | Release date: | 2001-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aminoglycoside binding displaces a divalent metal ion in a tRNA-neomycin B complex.
Nat.Struct.Biol., 8, 2001
|
|
2J4R
 
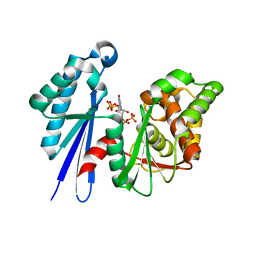 | | Structural Study of the Aquifex aeolicus PPX-GPPA enzyme | | Descriptor: | EXOPOLYPHOSPHATASE, GUANOSINE-5',3'-TETRAPHOSPHATE | | Authors: | Kristensen, O. | | Deposit date: | 2006-09-05 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of the Ppx/Gppa Phosphatase from Aquifex Aeolicus in Complex with the Alarmone Ppgpp
J.Mol.Biol., 375, 2008
|
|
1LO5
 
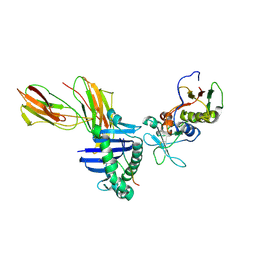 | | Crystal structure of the D227A variant of Staphylococcal enterotoxin A in complex with human MHC class II | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DR-1 beta chain, ... | | Authors: | Petersson, K, Thunnissen, M, Forsberg, G, Walse, B. | | Deposit date: | 2002-05-06 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of a SEA Variant in Complex with MHC Class II Reveals the Ability of SEA to Crosslink MHC Molecules
Structure, 10, 2002
|
|
1GZC
 
 | |
2IZ9
 
 | | MS2-RNA HAIRPIN (4ONE -5) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*U ONEP *AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Grahn, E, Stonehouse, N.J, Adams, C.J, Fridborg, K, Beigelman, L, Matulic-Adamic, J, Warriner, S.L, Stockley, P.G, Liljas, L. | | Deposit date: | 2006-07-25 | | Release date: | 2006-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Deletion of a Single Hydrogen Bonding Atom from the MS2 RNA Operator Leads to Dramatic Rearrangements at the RNA-Coat Protein Interface
Nucleic Acids Res., 28, 2000
|
|
2JFY
 
 | |
2JFP
 
 | |
2JAD
 
 | |
1H2X
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, Y473F MUTANT | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Fulop, V. | | Deposit date: | 2002-08-20 | | Release date: | 2002-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Electrostatic Effects and Binding Determinants in the Catalysis of Prolyl Oligopeptidase: Site Specific Mutagenesis at the Oxyanion Binding Site
J.Biol.Chem., 277, 2002
|
|
1H9Y
 
 | |
1GWU
 
 | |
1H9X
 
 | | Cytochrome cd1 Nitrite Reductase, reduced form | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CYTOCHROME CD1 NITRITE REDUCTASE, HEME C, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-03-23 | | Release date: | 2001-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of an Alternative Form of Paracoccus Pantotrophus Cytochrome Cd1 Nitrite Reductase
J.Biol.Chem., 276, 2001
|
|
1M61
 
 | |
