5CIE
 
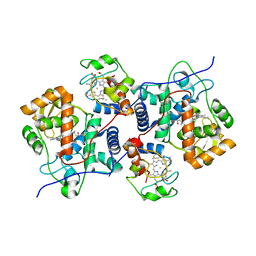 | |
5CIF
 
 | |
6MBI
 
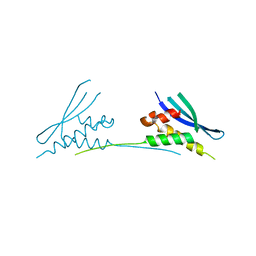 | |
5A4I
 
 | | The mechanism of Hydrogen activation by NiFE-hydrogenases | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.C, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
6LXG
 
 | | NMR solution structure of regulatory ACT domain of the Mycobacterium tuberculosis Rel protein | | Descriptor: | GTP pyrophosphokinase | | Authors: | Shin, J, Singal, B, Manimekalai, M.S.S, Gruber, G. | | Deposit date: | 2020-02-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Atomic structure of, and valine binding to the regulatory ACT domain of the Mycobacterium tuberculosis Rel protein.
Febs J., 288, 2021
|
|
5D82
 
 | |
7KP1
 
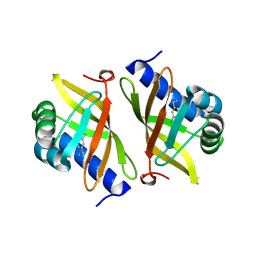
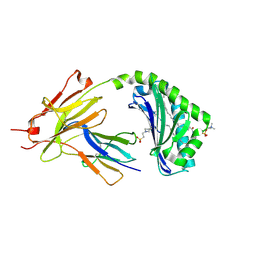 | | CD1a-42:2 SM binary complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, T-cell surface glycoprotein CD1a, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | CD1a selectively captures endogenous cellular lipids that broadly block T cell response.
J.Exp.Med., 218, 2021
|
|
5D81
 
 | |
7KP0
 
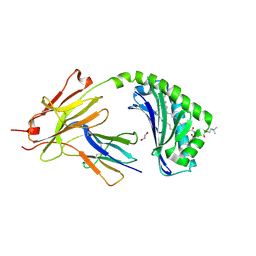 | | CD1a-42:1 SM binary complex | | Descriptor: | (4R,7S)-4-hydroxy-7-[(1S,2E)-1-hydroxyhexadec-2-en-1-yl]-N,N,N-trimethyl-4,9-dioxo-3,5-dioxa-8-aza-4lambda~5~-phosphadotriacontan-1-aminium, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD1a selectively captures endogenous cellular lipids that broadly block T cell response.
J.Exp.Med., 218, 2021
|
|
7KOZ
 
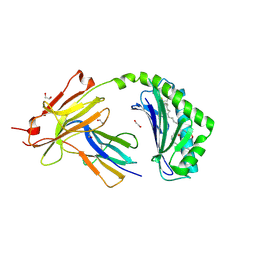 | | CD1a-36:2 SM binary complex | | Descriptor: | (4S,7S,17Z)-4-hydroxy-7-[(1S,2E)-1-hydroxyhexadec-2-en-1-yl]-N,N,N-trimethyl-4,9-dioxo-3,5-dioxa-8-aza-4lambda~5~-phosphahexacos-17-en-1-aminium, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CD1a selectively captures endogenous cellular lipids that broadly block T cell response.
J.Exp.Med., 218, 2021
|
|
5D83
 
 | |
5DAA
 
 | |
5BUN
 
 | | Crystal structure of an antigenic outer membrane protein ST50 from Salmonella Typhi | | Descriptor: | Outer membrane protein, octyl beta-D-glucopyranoside | | Authors: | Yoshimura, M, Chuankhayan, P, Lin, C.C, Chen, N.C, Yang, M.C, Fun, H.K. | | Deposit date: | 2015-06-04 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structure of an antigenic outer-membrane protein from Salmonella Typhi suggests a potential antigenic loop and an efflux mechanism.
Sci Rep, 5, 2015
|
|
5DUJ
 
 | | Crystal structure of ldtMt2 in complex with Faropenem adduct | | Descriptor: | (2R,5R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydro-1,3-thiazole-4-carboxylic acid, BETA-MERCAPTOETHANOL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5ANS
 
 | | Potent and selective inhibitors of MTH1 probe its role in cancer cell survival | | Descriptor: | 1-[4-amino-2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-1-yl]-2-methylpropan-2-ol, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE | | Authors: | Kettle, J.G, Alwan, H, Bista, M, Breed, J, Kack, H, Eckersley, K, Foote, K.M, Fillery, S, Goodwin, L, Jones, D, Lau, A, Nissink, J.W.M, Read, J, Scott, J, Taylor, B, Walker, G, Wissler, L. | | Deposit date: | 2015-09-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potent and Selective Inhibitors of Mth1 Probe its Role in Cancer Cell Survival.
J.Med.Chem., 59, 2016
|
|
5ANW
 
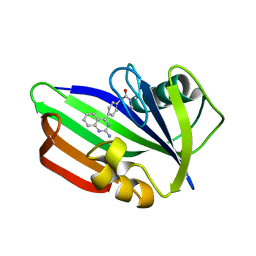 | | MTH1 in complex with compound 24 | | Descriptor: | 2-[4-(2-AMINOQUINAZOLIN-4-YL)PHENYL]-N,N-DIMETHYL-ACETAMIDE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2015-09-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Potent and Selective Inhibitors of Mth1 Probe its Role in Cancer Cell Survival.
J.Med.Chem., 59, 2016
|
|
5ANU
 
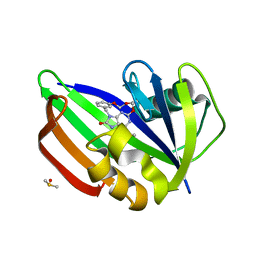 | | MTH1 in complex with compound 15 | | Descriptor: | 13-(METHYLAMINO)-23,24,25-TRIOXA-17,18,19,21-TETRAZATETRACYCLO-TRICOSA-1(3),2(10),4(11),12(14),13(18),16(19)-HEXAN-15-ONE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, DIMETHYL SULFOXIDE | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2015-09-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and Selective Inhibitors of Mth1 Probe its Role in Cancer Cell Survival.
J.Med.Chem., 59, 2016
|
|
5E6F
 
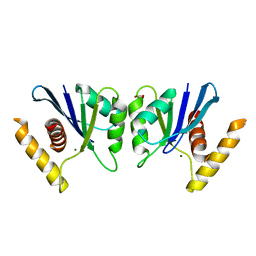 | | Canarypox virus resolvase | | Descriptor: | CNPV261 Holliday junction resolvase protein, D(-)-TARTARIC ACID, MAGNESIUM ION | | Authors: | Li, H, Hwang, Y, Perry, K, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Metal Binding Properties of a Poxvirus Resolvase.
J.Biol.Chem., 291, 2016
|
|
5E7J
 
 | |
5DU7
 
 | |
5E1I
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T210 | | Descriptor: | (2S,3R,4R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-(methylsulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, GLYCEROL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2015-09-29 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5BU4
 
 | | RIBONUCLEASE T1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-15 | | Release date: | 1998-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
7KOD
 
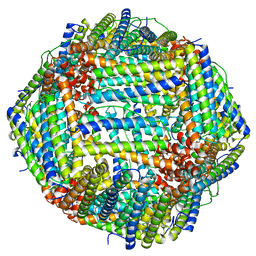 | | Cryo-EM structure of heavy chain mouse apoferritin | | Descriptor: | Ferritin heavy chain | | Authors: | Sun, M, Azumaya, C, Tse, E, Frost, A, Southworth, D, Verba, K.A, Cheng, Y, Agard, D.A. | | Deposit date: | 2020-11-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.655 Å) | | Cite: | Practical considerations for using K3 cameras in CDS mode for high-resolution and high-throughput single particle cryo-EM.
J.Struct.Biol., 213, 2021
|
|
6NKL
 
 | | 2.2 A resolution structure of VapBC-1 from nontypeable Haemophilus influenzae | | Descriptor: | Antitoxin VapB1, Ribonuclease VapC | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Molinaro, A.L, Daines, D.A. | | Deposit date: | 2019-01-07 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of VapBC-1 from Nontypeable Haemophilus influenzae and the Effect of PIN Domain Mutations on Survival during Infection.
J.Bacteriol., 201, 2019
|
|
5DZJ
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T206 in conformation A | | Descriptor: | (2~{R},3~{R},4~{R})-4-methyl-3-(2-oxidanylidene-2-propoxy-ethyl)sulfanyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, L,D-transpeptidase 2 | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
